

Identification and Expression Analysis of DNA Methyltransferase in Begonia masoniana
Received date: 2024-01-22
Accepted date: 2024-05-07
Online published: 2024-05-15
INTRODUCTION DNA methylation is one of the important epigenetic modifications involved in the regulation of plant genome stability, development and stress responses. DNA methylation introduces methylation groups into DNA molecules, thereby altering the activity of DNA segments. DNA methylation is catalyzed by DNA methyltransferase, a process by which methyl groups formed from S-adenosyl-L-methionine are transferred via covalent links to specific locations in the DNA sequence to form N4-methylcytosine, 5-methylcytosine, N6-methyladenine, or 7-methylguanine. However, there are few reports about the effects of DNA methyltransferase on leaf variegation formation and stress response of Begonia.
RATIONALE Studies have shown that DNA methylation is involved in regulating the formation of leaf color, flower color and leaf variegation, as well as responses to stresses and hormones. As an endemic species of Begonia, Begonia masoniana has unique and beautiful leaf markings, pink, dark green and light green in different developmental stages. It has high ornamental value and is an excellent foliage plant. Therefore, based on the genomic data, this study conducted genome-wide identification and expression pattern analysis of DNA methyltransferase genes, aiming to explore the genetic resources that regulate the formation of leaf variegation.
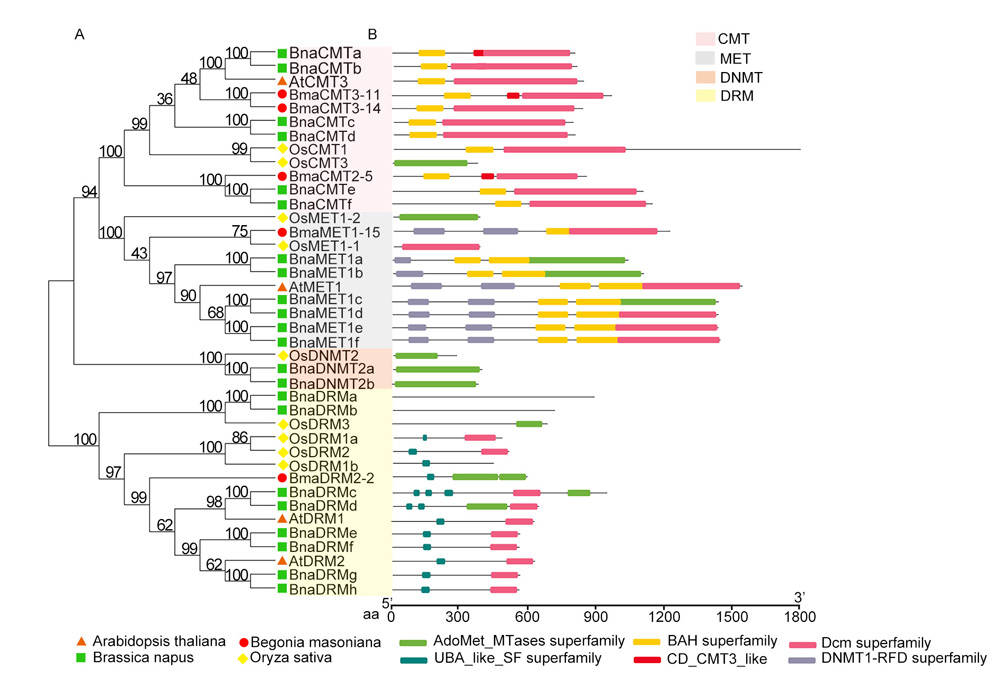
RESULTS To investigate whether DNA methyltransferase is involved in the regulation of leaf variegation formation and stress response in B. masoniana, bioinformatics analysis was used to identify the genes encoding DNA methyltransferase. Five genes were obtained from the genome of B. masoniana. According to the protein structural characteristics, their encoded proteins were divided into three categories including CMT, MET and DRM. The sequence length and intron number of these genes were significantly categorized into different subgroups, but their structure and conserved domains in the same subgroup were highly conserved. In addition, all the encoded proteins were predicted to locate in the nucleus. The promoters of these genes contain a large number of cis-acting elements such as light response, MYB binding, and plant hormone response elements. Analysis of hormone response patterns showed that the gene expression of CMT3 was significantly decreased under GA, SA and NAA, and the gene expression of CMT2 was significantly decreased under MeJA and NAA, while MET-type and DRM-type genes displayed significantly increased expression under GA and ABA treatments. In addition, tissue specific analysis showed that the expression levels of BmaCMT2-5 and BmaDRM2-2 in leaves were significantly higher than those of other tissues, while the expressions of these two genes and BmaMET1-15 in red part of leaves were significantly higher than that of green part, implying that these three genes may be involved in regulating the formation of leaf variegation.
CONCLUSION The structure and function of DNA methyltransferase genes vary significantly across different categories in B. masoniana. However, within each category, members display high conservation in gene structure, conserved domains, motifs, and evolutionary patterns. These genes are likely to play crucial roles in the growth and development of diverse tissues and organs, as well as in responding to various biological and abiotic stresses. Moreover, based on the differential expression patterns of BmaCMT2-5, BmaMET1-15, and BmaDRM2-2 genes between leaf variegation and non-variegation areas, coupled with the abundance of MYB regulatory elements related to anthocyanin synthesis in their promoters, it is hypothesized that these genes may contribute to the formation of leaf variegation. As the current understanding of the functional roles of these methyltransferase genes is largely speculative, future research should focus on their functional validation, which will involve utilizing reverse genetics techniques coupled with phenotypic observations to determine their involvement in specific biological processes. Additionally, physiological, biochemical, and molecular biological methods should be employed to elucidate the precise mechanisms of their actions.
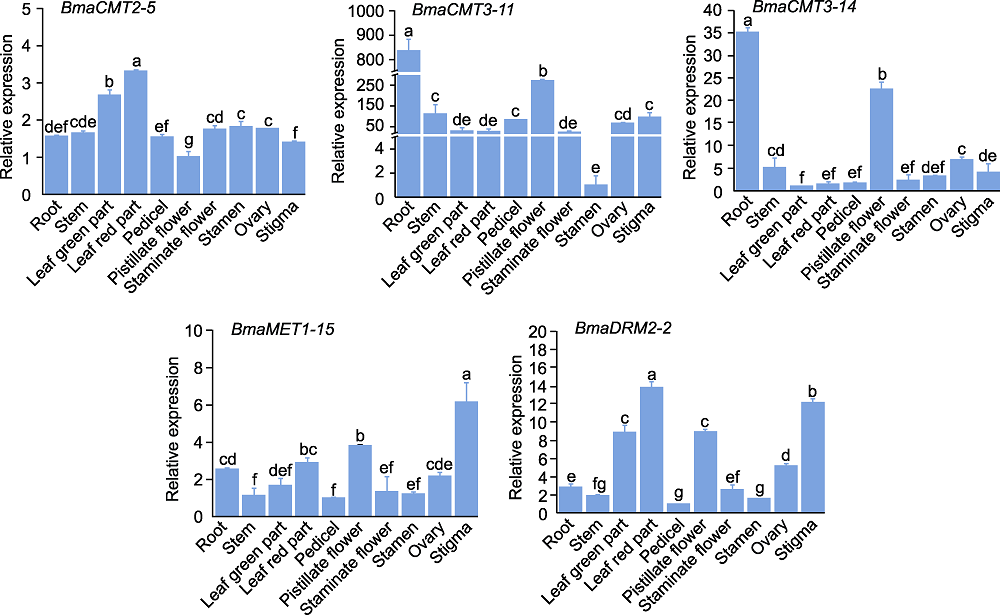
Relative expression levels of DNA methyltransferase genes in different tissues and organs of Begonia masoniana
Key words: Begonia masoniana; leaf variegation; DNA methyltransferase; phytohormones
Tingxin Chen , Min Fu , Na Li , Leilei Yang , Lingfei Li , Chunmei Zhong . Identification and Expression Analysis of DNA Methyltransferase in Begonia masoniana [J]. Chinese Bulletin of Botany, 2024 , 59(5) : 726 -737 . DOI: 10.11983/CBB24010
| [1] | 崔卫华, 管开云 (2013). 中国秋海棠属植物叶片斑纹多样性研究. 植物分类与资源学报 35, 119-127. |
| [2] | 戴若惠, 钱心妤, 孙静蕾, 芦涛, 贾绮玮, 陆天麒, 路梅, 饶玉春 (2023). 水稻叶色调控机制及相关基因研究进展. 植物学报 58, 799-812. |
| [3] | 杜文文, 段青, 马璐琳, 瞿素萍, 贾文杰, 王祥宁, 崔光芬 (2018). 7种秋海棠叶片斑纹结构及遗传特性分析. 西北植物学报 38, 2045-2052. |
| [4] | 关峰, 韦正乙, 王云鹏, 林春晶, 邢少辰, 马景勇 (2011). 内含子数量改变GUS基因的瞬时表达调控. 基因组学与应用生物学 30, 571-576. |
| [5] | 黎家, 李传友 (2019). 新中国成立70年来植物激素研究进展. 中国科学: 生命科学 49, 1227-1281. |
| [6] | 李景秀, 管开云, 孔繁才, 李爱荣 (2021). 中国秋海棠属植物资源概述. 中国野生植物资源 40(12), 35-44. |
| [7] | 李立奇, 万瑛 (2009). 蛋白质的亚细胞定位预测研究进展. 免疫学杂志 25, 602-604. |
| [8] | 冉浩然, 张毓, 陈简村, 于超, 张启翔, 罗乐 (2024). 观赏植物叶斑的研究进展. 植物遗传资源学报 25, 704-717. |
| [9] | 夏晗, 刘美芹, 尹伟伦, 卢存福, 夏新莉 (2008). 植物DNA甲基化调控因子研究进展. 遗传 30, 426-432. |
| [10] | 杨凯如, 贾绮玮, 金佳怡, 叶涵斐, 王盛, 陈芊羽, 管易安, 潘晨阳, 辛德东, 方媛, 王跃星, 饶玉春 (2022). 水稻黄绿叶调控基因YGL18的克隆与功能解析. 植物学报 57, 276- 287. |
| [11] | 杨婷, 薛珍珍, 李娜, 郎校安, 李凌飞, 钟春梅 (2021). 铁十字秋海棠斑叶发育过程内参基因筛选及验证. 园艺学报 48, 2251-2261. |
| [12] | 周陈平, 杨敏, 郭金菊, 邝瑞彬, 杨护, 黄炳雄, 魏岳荣 (2022). 番木瓜成熟过程中全基因组DNA甲基化和转录组变化分析. 园艺学报 49, 519-532. |
| [13] | Ahmad F, Huang X, Lan HX, Huma T, Bao YM, Huang J, Zhang HS (2014). Comprehensive gene expression analysis of the DNA (cytosine-5) methyltransferase family in rice (Oryza sativa L.). Genet Mol Res 13, 5159-5172. |
| [14] | Bennett M, Cleaves K, Hewezi T (2021). Expression patterns of DNA methylation and demethylation genes during plant development and in response to phytohormones. Int J Mol Sci 22, 9681. |
| [15] | Cao DY, Ju Z, Gao C, Mei XH, Fu DQ, Zhu HL, Luo YB, Zhu BZ (2014). Genome-wide identification of cytosine-5 DNA methyltransferases and demethylases in Solanum lycopersicum. Gene 550, 230-237. |
| [16] | Carey NS, Krogan NT (2017). The role of AUXIN RESPONSE FACTORs in the development and differential growth of inflorescence stems. Plant Signal Behav 12, e130-7492. |
| [17] | Chiu LW, Li L (2012). Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta 236, 1153-1164. |
| [18] | Fan SH, Liu HF, Liu J, Hua W, Xu SM, Li J (2020). Systematic analysis of the DNA methylase and demethylase gene families in rapeseed (Brassica napus L.) and their expression variations after salt and heat stresses. Int J Mol Sci 21, 953. |
| [19] | Furner IJ, Matzke M (2011). Methylation and demethylation of the Arabidopsis genome. Curr Opin Plant Biol 14, 137-141. |
| [20] | Gahlaut V, Samtani H, Khurana P (2020). Genome-wide identification and expression profiling of cytosine-5 DNA methyltransferases during drought and heat stress in wheat (Triticum aestivum). Genomics 112, 4796-4807. |
| [21] | Gianoglio S, Moglia A, Acquadro A, Comino C, Portis E (2017). The genome-wide identification and transcriptional levels of DNA methyltransferases and demethylases in globe artichoke. PLoS One 12, e0181669. |
| [22] | Gu TT, Ren S, Wang YH, Han YH, Li Y (2016). Characterization of DNA methyltransferase and demethylase genes in Fragaria vesca. Mol Genet Genomics 291, 1333- 1345. |
| [23] | Hong X, Scofield DG, Lynch M (2006). Intron size, abundance, and distribution within untranslated regions of genes. Mol Biol Evol 23, 2392-2404. |
| [24] | Jiang S, Guo YC (2020). Epigenetic clock: DNA methylation in aging. Stem Cells Int 2020, 1047896. |
| [25] | Li LF, Chen XL, Fang DM, Dong SS, Guo X, Li N, Campos-Dominguez L, Wang WG, Liu Y, Lang XA, Peng Y, Tian DK, Thomas DC, Mu WX, Liu M, Wu CY, Yang T, Zhang SZ, Yang LL, Yang JF, Liu ZJ, Zhang LS, Zhang XT, Chen F, Jiao YN, Guo YL, Hughes M, Wang W, Liu XF, Zhong CM, Li AR, Sahu SK, Yang HM, Wu E, Sharbrough J, Lisby M, Liu X, Xu X, Soltis DE, Van de Peer Y, Kidner C, Zhang SZ, Liu H (2022). Genomes shed light on the evolution of Begonia, a mega-diverse genus. New Phytol 234, 295-310. |
| [26] | Ma J, Li Q, Zhang L, Cai S, Liu YY, Lin JC, Huang RF, Yu YQ, Wen MZ, Xu TD (2022). High auxin stimulates callus through SDG8-mediated histone H3K36 methylation in Arabidopsis. J Integr Plant Biol 64, 2425-2437. |
| [27] | Parra G, Bradnam K, Rose AB, Korf I (2011). Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res 39, 5328-5337. |
| [28] | Parrilla-Doblas JT, Roldán-Arjona T, Ariza RR, Córdoba- Cañero D (2019). Active DNA demethylation in plants. Int J Mol Sci 20, 4683. |
| [29] | Tian J, Li KT, Zhang S, Zhang J, Song TT, Zhu YJ, Yao YC (2017). The structure and methylation level of the McMYB10 promoter determine the leaf color of Malus crabapple. HortScience 52, 520-526. |
| [30] | Wang QM, Wang L, Zhou YB, Cui JG, Wang YZ, Zhao CM (2016). Leaf patterning of Clivia miniata var. variegata is associated with differential DNA methylation. Plant Cell Rep 35, 167-184. |
| [31] | Wang Y, Liu S, Tian X, Fu Y, Jiang X, Li Y, Wang G (2018). Influence of light intensity on chloroplast development and pigment accumulation in wild-type and etiolated mutant plants of Anthurium andraeanum ‘Sonate’. Plant Signal Behav 13, e1482174. |
| [32] | Wu XX, Zhou Y, Yao D, Iqbal S, Gao ZH, Zhang Z (2020). DNA methylation of LDOX gene contributes to the floral colour variegation in peach. J Plant Physiol 246-247, 153116. |
| [33] | Xiao K, Chen J, He QXM, Wang YX, Shen HL, Sun L (2020). DNA methylation is involved in the regulation of pepper fruit ripening and interacts with phytohormones. J Exp Bot 71, 1928-1942. |
| [34] | Xu P, Su H, Jin R, Mao YX, Xu AN, Cheng HY, Wang YF, Meng Q (2020). Shading effects on leaf color conversion and biosynthesis of the major secondary metabolites in the albino tea cultivar ‘Yujinxiang’. J Agric Food Chem 68, 2528-2538. |
| [35] | Yan M, Yan Y, Wang P, Wang YP, Piao XM, Di P, Yang DC (2023). Genome-wide identification and expression analysis of auxin response factor (ARF) gene family in Panax ginseng indicates its possible roles in root development. Plants 12, 3943. |
| [36] | Yao MQ, Chen WW, Kong JH, Zhang XL, Shi NN, Zhong SL, Ma P, Gallusci P, Jackson S, Liu YL, Hong YG (2020). METHYLTRANSFERASE1 and ripening modulate vivipary during tomato fruit development. Plant Physiol 183, 1883-1897. |
| [37] | Yu H, Cui N, Guo K, Xu W, Wang HF (2023). Epigenetic changes in the regulation of carotenoid metabolism during honeysuckle flower development. Hortic Plant J 9, 577- 588. |
| [38] | Yue PT, Lu Q, Liu Z, Lv TX, Li XY, Bu HD, Liu WT, Xu YX, Yuan H, Wang AD (2020). Auxin-activated MdARF5 induces the expression of ethylene biosynthetic genes to initiate apple fruit ripening. New Phytol 226, 1781-1795. |
| [39] | Zhang Y, Zhao GY, Li YS, Mo N, Zhang J, Liang Y (2017). Transcriptomic analysis implies that GA regulates sex expression via ethylene-dependent and ethylene-indepen- dent pathways in cucumber (Cucumis sativus L.). Front Plant Sci 8, 10. |
| [40] | Zheng J, Wu H, Zhu HB, Huang CY, Liu C, Chang YS, Kong ZC, Zhou ZH, Wang GW, Lin YJ, Chen H (2019). Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol 223, 705-721. |
/
| 〈 |
|
〉 |