

Indispensable Material for Germination: Long-lived mRNAs of Plant Seed
Received date: 2024-01-10
Accepted date: 2024-03-30
Online published: 2024-05-08
Higher plants usually start from seed germination and re-form seeds after vegetative growth and reproductive development, thus completing the life cycle. Carbohydrates, lipids, proteins, mRNA and other macromolecular substances accumulated in seeds are crucial to maintain the germination potential of seeds, some of mRNA can be preserved for a long time without degradation, known as long-lived mRNA. In rice, long-lived mRNA associated with germination began to be transcribed and accumulated 10 to 20 days after flowering, and some long-lived mRNA associated with dormancy and stress response were transcribed and preserved in cells from 20 days after flowering to seed maturity. There are many kinds of long-lived mRNA, mainly including some protein synthesis mRNA, energy metabolism mRNA, cytoskeleton mRNA and some stress response related mRNA, such as small heat shock protein, LEA (late embryogenesis abundant) family proteins. Transcriptomic analysis found that the promoter regions of many genes contain ABA- or GA-associated cis-acting elements, and there are about 500 differentially expressed long-lived mRNAs in the Arabidopsis atabi5 (ABA-insensitive 5) mutant seeds that differ from the wild type, suggesting that abscisic acid (ABA) and gibberellin (GA) are the key hormones that influence the type of long-lived mRNA. Long-lived mRNAs are usually cross-linked with a single ribosome, RNA binding protein, which exists in cells in the form of P-bodies (PBs) to protect the mRNA from degradation. However, long-lived mRNAs associated with seed dormancy are gradually degraded during seed post-ripening, and the oxidative modification of some specific long-lived mRNAs is also a biological phenomenon to break seed dormancy. During the long-term storage of seeds, the random degradation of long-lived mRNA is directly related to the life and vitality of seeds, and the retained mRNA is translated into protein to help the rapid germination of seeds in the early stage of imbibition. In this paper, the characteristics and functions of long-lived mRNA are reviewed, and some future scientific issues are discussed to provide a reference for further understanding of the molecular mechanisms of seed dormancy, germination and longevity.
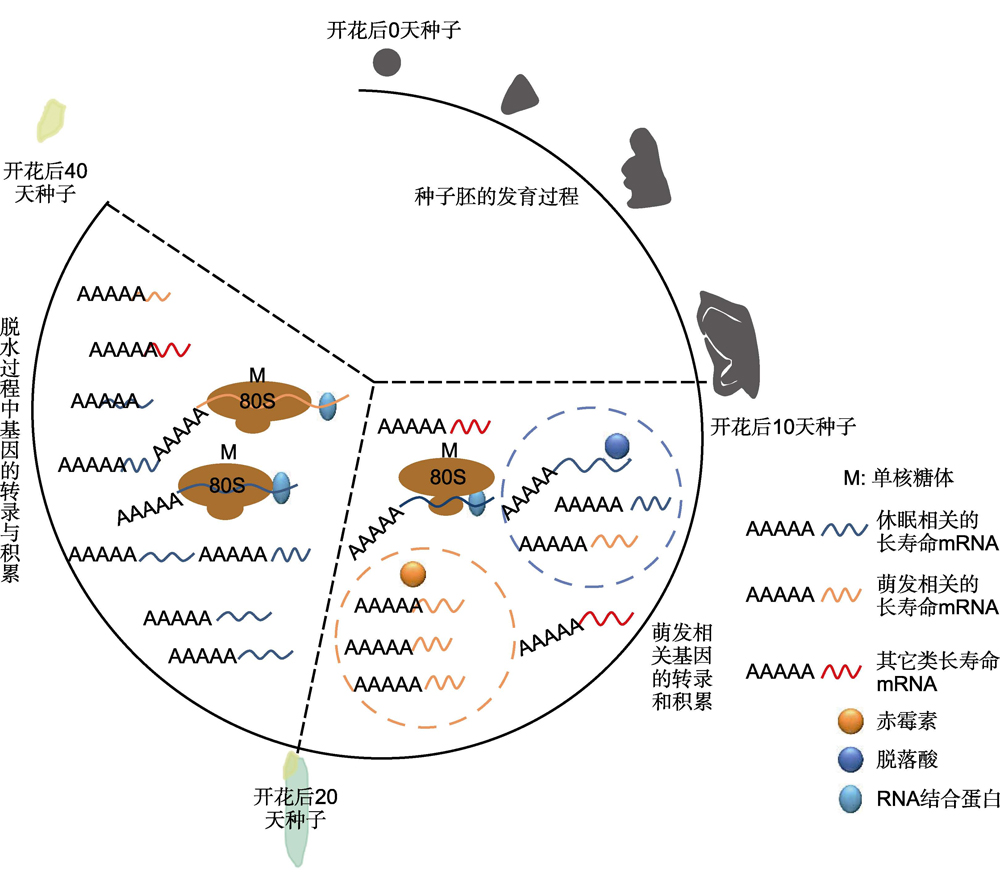
Key words: long-lived mRNAs; seed dormancy; seed germination; seed storage
Xiaobo Zhu , Zhang Dong , Mengjin Zhu , Jin Hu , Cheng Lin , Min Chen , Yajing Guan . Indispensable Material for Germination: Long-lived mRNAs of Plant Seed[J]. Chinese Bulletin of Botany, 2024 , 59(3) : 355 -372 . DOI: 10.11983/CBB24006
| [1] | Aalen RB, Salehian Z, Steinum TM (2001). Stability of barley aleurone transcripts: dependence on protein synthesis, influence of the starchy endosperm and destabilization by GA3. Physiol Plant 112, 403-413. |
| [2] | Almoguera C, Jordano J (1992). Developmental and environmental concurrent expression of sunflower dry-seed- stored low-molecular-weight heat-shock protein and Lea mRNAs. Plant Mol Biol 19, 781-792. |
| [3] | Ariizumi T, Hauvermale AL, Nelson SK, Hanada A, Yamaguchi S, Steber CM (2013). Lifting DELLA repression of Arabidopsis seed germination by nonproteolytic gibberellin signaling. Plant Physiol 162, 2125-2139. |
| [4] | Ariizumi T, Steber CM (2007). Seed germination of GA- insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 19, 791-804. |
| [5] | Aspart L, Meyer Y, Laroche M, Penon P (1984). Developmental regulation of the synthesis of proteins encoded by stored mRNA in radish embryos. Plant Physiol 76, 664-673. |
| [6] | Bazin J, Langlade N, Vincourt P, Arribat S, Balzergue S, El-Maarouf-Bouteau H, Bailly C (2011). Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell 23, 2196-2208. |
| [7] | Beltrán-Pe?a E, Ortíz-López A, de Jiménez ES (1995). Synthesis of ribosomal proteins from stored mRNAs early in seed germination. Plant Mol Biol 28, 327-336. |
| [8] | Bentsink L, Jowett J, Hanhart CJ, Koornneef M (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA 103, 17042-17047. |
| [9] | Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006). Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46, 805-822. |
| [10] | Chai MF, Queralta Castillo I, Sonntag A, Wang SX, Zhao ZL, Liu W, Du J, Xie HL, Liao FQ, Yun JF, Jiang QZ, Sun J, Molina I, Wang ZY (2021). A seed coat-specific β-ketoacyl-CoA synthase, KCS12, is critical for preserving seed physical dormancy. Plant Physiol 186, 1606-1615. |
| [11] | Chai MF, Zhou C, Molina I, Fu CX, Nakashima J, Li GF, Zhang WZ, Park J, Tang YH, Jiang QZ, Wang ZY (2016). A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc Natl Acad Sci USA 113, 6997-7002. |
| [12] | Chen FY, Li Y, Li XY, Li WL, Xu JM, Cao H, Wang Z, Li Y, Soppe WJJ, Liu YX (2021). Ectopic expression of the Arabidopsis florigen gene FLOWERING LOCUS T in seeds enhances seed dormancy via the GA and DOG1 pathways. Plant J 107, 909-924. |
| [13] | Chen HH, Ruan JX, Chu P, Fu W, Liang ZW, Li Y, Tong JH, Xiao LT, Liu J, Li CL, Huang SZ (2020a). AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant J 101, 310-323. |
| [14] | Chen HH, Tong JH, Fu W, Liang ZW, Ruan JX, Yu YG, Song X, Yuan LB, Xiao LT, Liu J, Cui YH, Huang SZ, Li CL (2020b). The H3K27me3 demethylase RELATIVE OF EARLY FLOWERING6 suppresses seed dormancy by inducing abscisic acid catabolism. Plant Physiol 184, 1969-1978. |
| [15] | Chen KG, An YQC (2006). Transcriptional responses to gibberellin and abscisic acid in barley aleurone. J Integr Plant Biol 48, 591-612. |
| [16] | Chen Y, Xiang ZP, Liu M, Wang SY, Zhang L, Cai D, Huang Y, Mao DD, Fu J, Chen LB (2023). ABA biosynthesis gene OsNCED3 contributes to preharvest sprouting resistance and grain development in rice. Plant Cell Environ 46, 1384-1401. |
| [17] | Chong SN, Ravindran P, Kumar PP (2022). Regulation of primary seed dormancy by MAJOR LATEX PROTEIN- LIKE PROTEIN329 in Arabidopsis is dependent on DNA-BINDING ONE ZINC FINGER6. J Exp Bot 73, 6838-6852. |
| [18] | De Guzman MK, Parween S, Butardo VM, Alhambra CM, Anacleto R, Seiler C, Bird AR, Chow CP, Sreenivasulu N (2017). Investigating glycemic potential of rice by unraveling compositional variations in mature grain and starch mobilization patterns during seed germination. Sci Rep 7, 5854. |
| [19] | Deng GL, Sun HQ, Hu YL, Yang YR, Li P, Chen YL, Zhu Y, Zhou Y, Huang JL, Neill SJ, Hu XY (2023). A transcription factor WRKY36 interacts with AFP2 to break primary seed dormancy by progressively silencing DOG1 in Arabidopsis. New Phytol 238, 688-704. |
| [20] | Ding ZJ, Yan JY, Li GX, Wu ZC, Zhang SQ, Zheng SJ (2014). WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J 79, 810-823. |
| [21] | Dure L, Waters L (1965). Long-lived messenger RNA: evidence from cotton seed germination. Science 147, 410-412. |
| [22] | Fleming MB, Patterson EL, Reeves PA, Richards CM, Gaines TA, Walters C (2018). Exploring the fate of mRNA in aging seeds: protection, destruction, or slow decay? J Exp Bot 69, 4309-4321. |
| [23] | Fu K, Song WH, Chen C, Mou CL, Huang YS, Zhang FL, Hao QX, Wang P, Ma TF, Chen YP, Zhu ZY, Zhang M, Tong QK, Liu X, Jiang L, Wan JM (2022). Improving pre-harvest sprouting resistance in rice by editing OsABA8ox using CRISPR/Cas9. Plant Cell Rep 41, 2107-2110. |
| [24] | Gao F, Rampitsch C, Chitnis VR, Humphreys GD, Jordan MC, Ayele BT (2013). Integrated analysis of seed proteome and mRNA oxidation reveals distinct post-transcriptional features regulating dormancy in wheat (Triticum aestivum L.). Plant Biotechnol J 11, 921-932. |
| [25] | Griffiths J, Barrero JM, Taylor J, Helliwell CA, Gubler F (2011). ALTERED MERISTEM PROGRAM 1 is involved in development of seed dormancy in Arabidopsis. PLoS One 6, e20408. |
| [26] | Guan YJ, Hu J (2000). Seed Science (Condensed edition). Beijing: China Agriculture Press. pp. 64. (in Chinese) |
| 关亚静, 胡晋 (2000). 种子学(精编版). 北京: 中国农业出版社. pp. 64. | |
| [27] | Guo NH, Tang SJ, Wang YK, Chen W, An RH, Ren ZL, Hu SK, Tang SQ, Wei XJ, Shao GN, Jiao GA, Xie LH, Wang L, Chen Y, Zhao FL, Sheng ZH, Hu PS (2024). A mediator of OsbZIP46 deactivation and degradation negatively regulates seed dormancy in rice. Nat Commun 15, 1134. |
| [28] | Harris B, Dure L 3rd (1978). Developmental regulation in cotton seed germination: polyadenylation of stored messenger RNA. Biochemistry 17, 3250-3256. |
| [29] | Hazra A, Varshney V, Verma P, Kamble NU, Ghosh S, Achary RK, Gautam S, Majee M (2022). Methionine sulfoxide reductase B5 plays a key role in preserving seed vigor and longevity in rice (Oryza sativa). New Phytol 236, 1042-1060. |
| [30] | He DL, Han C, Yao JL, Shen SH, Yang PF (2011). Constructing the metabolic and regulatory pathways in germinating rice seeds through proteomic approach. Proteomics 11, 2693-2713. |
| [31] | He WP, Wang R, Zhang Q, Fan MX, Lyu Y, Chen S, Chen DF, Chen XW (2023). E3 ligase ATL5 positively regulates seed longevity by mediating the degradation of ABT1 in Arabidopsis. New Phytol 239, 1754-1770. |
| [32] | Howell KA, Narsai R, Carroll A, Ivanova A, Lohse M, Usadel B, Millar AH, Whelan J (2009). Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol 149, 961-980. |
| [33] | Huang YS, Song JW, Hao QX, Mou CL, Wu HM, Zhang FL, Zhu ZY, Wang P, Ma TF, Fu K, Chen YP, Nguyen T, Liu SJ, Jiang L, Wan JM (2023). WEAK SEED DORMANCY 1, an aminotransferase protein, regulates seed dormancy in rice through the GA and ABA pathways. Plant Physiol Biochem 202, 107923. |
| [34] | Huh JH, Bauer MJ, Hsieh TF, Fischer R (2007). Endosperm gene imprinting and seed development. Curr Opin Genet Dev 17, 480-485. |
| [35] | Hundertmark M, Hincha DK (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9, 118. |
| [36] | Ingle J, Hageman RH (1965). Metabolic changes associated with the germination of corn. II. Nucleic acid metabolism. Plant Physiol 40, 48-53. |
| [37] | Ishibashi N, Yamauchi D, Minamikawa T (1990). Stored mRNA in cotyledons of Vigna unguiculata seeds: nucleotide sequence of cloned cDNA for a stored mRNA and induction of its synthesis by precocious germination. Plant Mol Biol 15, 59-64. |
| [38] | Isono K, Shimizu M, Yoshimoto K, Niwa Y, Satoh K, Yokota A, Kobayashi H (1997). Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domains of σ70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc Natl Acad Sci USA 94, 14948-14953. |
| [39] | Itoh JI, Nonomura KI, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol 46, 23-47. |
| [40] | Ivanov R, Tiedemann J, Czihal A, Baumlein H (2012). Transcriptional regulator AtET2 is required for the induction of dormancy during late seed development. J Plant Physiol 169, 501-508. |
| [41] | Iwasaki M, Hyv?rinen L, Piskurewicz U, Lopez-Molina L (2019). Non-canonical RNA-directed DNA methylation participates in maternal and environmental control of seed dormancy. eLife 8, e37434. |
| [42] | Jacobsen JV, Pearce DW, Poole AT, Pharis RP, Mander LN (2002). Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol Plant 115, 428-441. |
| [43] | Kearly A, Nelson ADL, Skirycz A, Chodasiewicz M (2024). Composition and function of stress granules and P-bodies in plants. Semin Cell Dev Biol 156, 167-175. |
| [44] | Kim S, Huh SM, Han HJ, Lee GS, Hwang YS, Cho MH, Kim BG, Song JS, Chung JH, Nam MH, Ji H, Kim KH, Yoon IS (2023). A rice seed-specific glycine-rich protein OsDOR1 interacts with GID1 to repress GA signaling and regulates seed dormancy. Plant Mol Biol 111, 523-539. |
| [45] | Kim W, Lee Y, Park J, Lee N, Choi G (2013). HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol 54, 555-572. |
| [46] | Kimura M, Nambara E (2010). Stored and neosynthesized mRNA in Arabidopsis seeds: effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition. Plant Mol Biol 73, 119-129. |
| [47] | Kong QM, Lin CLG (2010). Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell Mol Life Sci 67, 1817-1829. |
| [48] | Li XY, Chen TT, Li Y, Wang Z, Cao H, Chen FY, Li Y, Soppe WJJ, Li WL, Liu YX (2019). ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression. Plant Cell 31, 832-847. |
| [49] | Li Y, Chen FY, Yang Y, Han Y, Ren ZY, Li XY, Soppe WJJ, Cao H, Liu YX (2023). The Arabidopsis pre-mRNA 3' end processing related protein FIP1 promotes seed dormancy via the DOG1 and ABA pathways. Plant J 115, 494-509. |
| [50] | Liu SP, Li L, Wang WL, Xia GM, Liu SW (2024). TaSRO1 interacts with TaVP1 to modulate seed dormancy and pre-harvest sprouting resistance in wheat. J Integr Plant Biol 66, 36-53. |
| [51] | Liu SR, Yang LW, Li JL, Tang WJ, Li JG, Lin RC (2021). FHY3 interacts with phytochrome B and regulates seed dormancy and germination. Plant Physiol 187, 289-302. |
| [52] | Liu XL, Li N, Chen AY, Saleem N, Jia QL, Zhao CZ, Li WQ, Zhang M (2023). FUSCA3-induced AINTEGUMENTA-like 6 manages seed dormancy and lipid metabolism. Plant Physiol 193, 1091-1108. |
| [53] | Martinet W, De Meyer GRY, Herman AG, Kockx MM (2005). RNA damage in human atherosclerosis: pathophysiological significance and implications for gene expression studies. RNA Biol 2, 4-7. |
| [54] | Martínez-Andújar C, Ordiz MI, Huang ZL, Nonogaki M, Beachy RN, Nonogaki H (2011). Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc Natl Acad Sci USA 108, 17225-17229. |
| [55] | Masaki S, Yamada T, Hirasawa T, Todaka D, Kanekatsu M (2008). Proteomic analysis of RNA-binding proteins in dry seeds of rice after fractionation by ssDNA affinity column chromatography. Biotechnol Lett 30, 955-960. |
| [56] | Matilla AJ (2022). Exploring breakthroughs in three traits belonging to seed life. Plants (Basel) 11, 490. |
| [57] | Mertens J, Aliyu H, Cowan DA (2018). LEA proteins and the evolution of the WHy domain. Appl Environ Microbiol 84, e00539-18. |
| [58] | Montoya-García L, Mu?oz-Ocotero V, Aguilar R, Sánchez de Jiménez E (2002). Regulation of acidic ribosomal protein expression and phosphorylation in maize. Biochemistry 41, 10166-10172. |
| [59] | Müller K, Carstens AC, Linkies A, Torres MA, Leubner-Metzger G (2009). The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol 184, 885-897. |
| [60] | Nakabayashi K, Bartsch M, Xiang Y, Miatton E, Pellengahr S, Yano R, Seo M, Soppe WJJ (2012). The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24, 2826-2838. |
| [61] | Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41, 697-709. |
| [62] | Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Utsugi S, Ogawa T, Handa H, Ishida H, Mori M, Kawaura K, Ogihara Y, Miura H (2011). A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23, 3215-3229. |
| [63] | Narro-Diego L, López-González L, Jarillo JA, Pi?eiro M (2017). The PHD-containing protein EARLY BOLTING IN SHORT DAYS regulates seed dormancy in Arabidopsis. Plant Cell Environ 40, 2393-2405. |
| [64] | Narsai R, Howell KA, Millar AH, O'Toole N, Small I, Whelan J (2007). Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19, 3418-3436. |
| [65] | Nelson SK, Ariizumi T, Steber CM (2017). Biology in the dry seed: transcriptome changes associated with dry seed dormancy and dormancy loss in the Arabidopsis GA-insensitive sleepy1-2 mutant. Front Plant Sci 8, 2158. |
| [66] | Ni?oles R, Planes D, Arjona P, Ruiz-Pastor C, Chazarra R, Renard J, Bueso E, Forment J, Serrano R, Kranner I, Roach T, Gadea J (2022). Comparative analysis of wild-type accessions reveals novel determinants of Arabidopsis seed longevity. Plant Cell Environ 45, 2708-2728. |
| [67] | Nomura T, Ueno M, Yamada Y, Takatsuto S, Takeuchi Y, Yokota T (2007). Roles of brassinosteroids and related mRNAs in pea seed growth and germination. Plant Physiol 143, 1680-1688. |
| [68] | Okamoto M, Tatematsu K, Matsui A, Morosawa T, Ishida J, Tanaka M, Endo TA, Mochizuki Y, Toyoda T, Kamiya Y, Shinozaki K, Nambara E, Seki M (2010). Genome- wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J 62, 39-51. |
| [69] | Oracz K, El-Maarouf Bouteau H, Farrant JM, Cooper K, Belghazi M, Job C, Job D, Corbineau F, Bailly C (2007). ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J 50, 452-465. |
| [70] | Qin P, Zhang GH, Hu BH, Wu J, Chen WL, Ren ZJ, Liu YL, Xie J, Yuan H, Tu BT, Ma B, Wang YP, Ye LM, Li LG, Xiang CB, Li SG (2021). Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism. Sci Adv 7, eabc8873. |
| [71] | Rajjou L, Debeaujon I (2008). Seed longevity: survival and maintenance of high germination ability of dry seeds. C R Biol 331, 796-805. |
| [72] | Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004). The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134, 1598-1613. |
| [73] | Ravindran P, Verma V, Stamm P, Kumar PP (2017). A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol Plant 10, 1307-1320. |
| [74] | Rueda-Romero P, Barrero-Sicilia C, Gómez-Cadenas A, Carbonero P, O?ate-Sánchez L (2012). Arabidopsis thaliana DOF6 negatively affects germination in non-after- ripened seeds and interacts with TCP14. J Exp Bot 63, 1937-1949. |
| [75] | Sajeev N, Bai B, Bentsink L (2019). Seeds: a unique system to study translational regulation. Trends Plant Sci 24, 487-495. |
| [76] | Sajeev N, Baral A, America AHP, Willems LAJ, Merret R, Bentsink L (2022). The mRNA-binding proteome of a critical phase transition during Arabidopsis seed germination. New Phytol 233, 251-264. |
| [77] | Sánchez-de-Jiménez E, Aguilar R, Dinkova T (1997). S6 ribosomal protein phosphorylation and translation of stored mRNA in maize. Biochimie 79, 187-194. |
| [78] | Sano N, Masaki S, Tanabata T, Yamada T, Hirasawa T, Kashiwagi M, Kanekatsu M (2013). RNA-binding proteins associated with desiccation during seed development in rice. Biotechnol Lett 35, 1945-1952. |
| [79] | Sano N, Ono H, Murata K, Yamada T, Hirasawa T, Kanekatsu M (2015). Accumulation of long-lived mRNAs associated with germination in embryos during seed development of rice. J Exp Bot 66, 4035-4046. |
| [80] | Sano N, Permana H, Kumada R, Shinozaki Y, Tanabata T, Yamada T, Hirasawa T, Kanekatsu M (2012). Proteomic analysis of embryonic proteins synthesized from long- lived mRNAs during germination of rice seeds. Plant Cell Physiol 53, 687-698. |
| [81] | Sano N, Takebayashi Y, To A, Mhiri C, Rajjou L, Nakagami H, Kanekatsu M (2019). Shotgun proteomic analysis highlights the roles of long-lived mRNAs and de novo transcribed mRNAs in rice seeds upon imbibition. Plant Cell Physiol 60, 2584-2596. |
| [82] | Sharma SN, Maheshwari A, Sharma C, Shukla N (2018). Gene expression patterns regulating the seed metabolism in relation to deterioration/ageing of primed mung bean (Vigna radiata L.) seeds. Plant Physiol Biochem 124, 40-49. |
| [83] | Shen WZ, Yao X, Ye TT, Ma S, Liu X, Yin XM, Wu Y (2018). Arabidopsis aspartic protease ASPG1 affects seed dormancy, seed longevity and seed germination. Plant Cell Physiol 59, 1415-1431. |
| [84] | Shu K, Zhang HW, Wang SF, Chen ML, Wu YR, Tang SY, Liu CY, Feng YQ, Cao XF, Xie Q (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet 9, e1003577. |
| [85] | Siloto RMP, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006). The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18, 1961-1974. |
| [86] | Song WH, Hao QX, Cai MY, Wang YH, Zhu XJ, Liu X, Huang YS, Nguyen T, Yang CY, Yu JF, Wu HM, Chen LM, Tian YL, Jiang L, Wan JM (2020). Rice OsBT1 regulates seed dormancy through the glycometabolism pathway. Plant Physiol Biochem 151, 469-476. |
| [87] | Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, Stein N, Weschke W, Strickert M, Close TJ, Stitt M, Graner A, Wobus U (2008). Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146, 1738-1758. |
| [88] | Standart N, Weil D (2018). P-bodies: cytosolic droplets for coordinated mRNA storage. Trends Genet 34, 612-626. |
| [89] | Sugimoto M, Oono Y, Kawahara Y, Gusev O, Maekawa M, Matsumoto T, Levinskikh M, Sychev V, Novikova N, Grigoriev A (2016). Gene expression of rice seeds surviving 13- and 20-month exposure to space environment. Life Sci Space Res (Amst) 11, 10-17. |
| [90] | Suriyasak C, Oyama Y, Ishida T, Mashiguchi K, Yamaguchi S, Hamaoka N, Iwaya-Inoue M, Ishibashi Y (2020). Mechanism of delayed seed germination caused by high temperature during grain filling in rice (Oryza sativa L.). Sci Rep 10, 17378. |
| [91] | Suzuki Y, Minamikawa T (1985). On the role of stored mRNA in protein synthesis in embryonic axes of germinating Vigna unguiculata seeds. Plant Physiol 79, 327-331. |
| [92] | Taylor RE, Waterworth W, West CE, Foyer CH (2023). WHIRLY proteins maintain seed longevity by effects on seed oxygen signaling during imbibition. Biochem J 480, 941-956. |
| [93] | Tiller K, Eisermann A, Link G (1991). The chloroplast transcription apparatus from mustard (Sinapis alba L.). Evidence for three different transcription factors which resemble bacterial sigma factors. Eur J Biochem 198, 93-99. |
| [94] | Vaistij FE, Gan YB, Penfield S, Gilday AD, Dave A, He ZS, Josse EM, Choi G, Halliday KJ, Graham IA (2013). Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc Natl Acad Sci USA 110, 10866-10871. |
| [95] | Villa-Hernández JM, Dinkova TD, Aguilar-Caballero R, Rivera-Cabrera F, Sánchez de Jiménez E, Pérez-Flores LJ (2013). Regulation of ribosome biogenesis in maize embryonic axes during germination. Biochimie 95, 1871-1879. |
| [96] | Wang BQ, Wang SY, Tang YQ, Jiang LL, He W, Lin QL, Yu F, Wang L (2022). Transcriptome-wide characterization of seed aging in rice: identification of specific long-lived mRNAs for seed longevity. Front Plant Sci 13, 857390. |
| [97] | Wang CL, Lyu Y, Zhang Q, Guo HY, Chen DF, Chen XW (2023). Disruption of BG14 results in enhanced callose deposition in developing seeds and decreases seed longevity and seed dormancy in Arabidopsis. Plant J 113, 1080-1094. |
| [98] | Wang HT, Zhang YM, Xiao N, Zhang G, Wang F, Chen XY, Fang RX (2020a). Rice GERMIN-LIKE PROTEIN 2-1 functions in seed dormancy under the control of abscisic acid and gibberellic acid signaling pathways. Plant Physiol 183, 1157-1170. |
| [99] | Wang Q, Lin QB, Wu T, Duan EC, Huang YS, Yang CY, Mou CL, Lan J, Zhou CL, Xie K, Liu X, Zhang X, Guo XP, Wang J, Jiang L, Wan JM (2020b). OsDOG1L-3 regulates seed dormancy through the abscisic acid pathway in rice. Plant Sci 298, 110570. |
| [100] | Wang Z, Cao H, Sun YZ, Li XY, Chen FY, Carles A, Li Y, Ding M, Zhang C, Deng X, Soppe WJJ, Liu YX (2013). Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25, 149-166. |
| [101] | Wei J, Wu XT, Li XY, Soppe WJJ, Cao H, Liu YX (2023). Overexpression of Taetr1-1 promotes enhanced seed dormancy and ethylene insensitivity in wheat. Planta 258, 56. |
| [102] | Wise MJ (2003). LEAping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics 4, 52. |
| [103] | Wu J, Jin YJ, Liu C, Vonapartis E, Liang JH, Wu WJ, Gazzarrini S, He JN, Yi MF (2019). GhNAC83 inhibits corm dormancy release by regulating ABA signaling and cytokinin biosynthesis in Gladiolus hybridus. J Exp Bot 70, 1221-1237. |
| [104] | Xiang Y, Nakabayashi K, Ding J, He F, Bentsink L, Soppe WJJ (2014). REDUCED DORMANCY5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 26, 4362-4375. |
| [105] | Xu F, Tang JY, Wang SX, Cheng X, Wang HR, Ou SJ, Gao SP, Li BS, Qian YW, Gao CX, Chu CC (2022). Antagonistic control of seed dormancy in rice by two bHLH transcription factors. Nat Genet 54, 1972-1982. |
| [106] | Yazdanpanah F, Willems LAJ, He HZ, Hilhorst HWM, Bentsink L (2022). A role for allantoate amidohydrolase (AtAAH) in the germination of Arabidopsis thaliana seeds. Plant Cell Physiol 63, 1298-1308. |
| [107] | Ye H, Feng JH, Zhang LH, Zhang JF, Mispan MS, Cao ZQ, Beighley DH, Yang JC, Gu XY (2015). Map-based cloning of seed Dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol 169, 2152-2165. |
| [108] | Yoshiyama K, Conklin PA, Huefner ND, Britt AB (2009). Suppressor of gamma response 1 ( SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci USA 106, 12843-12848. |
| [109] | Zhang Q, Pritchard J, Mieog J, Byrne K, Colgrave ML, Wang JR, Ral JPF (2021). Overexpression of a wheat α-amylase type 2 impact on starch metabolism and abscisic acid sensitivity during grain germination. Plant J 108, 378-393. |
| [110] | Zhao FK, Ma Q, Li YJ, Jiang MH, Zhou ZJ, Meng S, Peng Y, Zhang JH, Ye NH, Liu BH (2023). OsNAC2 regulates seed dormancy and germination in rice by inhibiting ABA catabolism. Biochem Biophys Res Commun 682, 335-342. |
| [111] | Zhao L, Wang H, Fu YB (2020a). Analysis of stored mRNA degradation in acceleratedly aged seeds of wheat and canola in comparison to Arabidopsis. Plants (Basel) 9, 1707. |
| [112] | Zhao L, Wang S, Fu YB, Wang H (2020b). Arabidopsis seed stored mRNAs are degraded constantly over aging time, as revealed by new quantification methods. Front Plant Sci 10, 1764. |
| [113] | Zheng J, Chen FY, Wang Z, Cao H, Li XY, Deng X, Soppe WJJ, Li Y, Liu YX (2012). A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol 193, 605-616. |
| [114] | Zheng LP, Otani M, Kanno Y, Seo M, Yoshitake Y, Yoshimoto K, Sugimoto K, Kawakami N (2022). Seed dormancy 4 like1 of Arabidopsis is a key regulator of phase transition from embryo to vegetative development. Plant J 112, 460-475. |
| [115] | Zhu CC, Wang CX, Lu CY, Wang JD, Zhou Y, Xiong M, Zhang CQ, Liu QQ, Li QF (2021). Genome-wide identification and expression analysis of OsbZIP09 target genes in rice reveal its mechanism of controlling seed germination. Int J Mol Sci 22, 1661. |
| [116] | Zhu HF, Xie WX, Xu DC, Miki D, Tang K, Huang CF, Zhu JK (2018). DNA demethylase ROS1 negatively regulates the imprinting of DOGL4 and seed dormancy in Arabidopsis thaliana. Proc Natl Acad Sci USA 115, E9962- E9970. |
| [117] | Zinsmeister J, Berriri S, Basso DP, Ly-Vu B, Dang TT, Lalanne D, da Silva EAA, Leprince O, Buitink J (2020). The seed-specific heat shock factor A9 regulates the depth of dormancy in Medicago truncatula seeds via ABA signaling. Plant Cell Environ 43, 2508-2522. |
/
| 〈 |
|
〉 |