

Identification of Sex Determination Molecular Marker Based on Genome-wide Association Study of Idesia polycarpa
Received date: 2024-02-28
Accepted date: 2024-05-06
Online published: 2024-05-07
RATIONALE In plants, the origin and evolution of sex chromosomes was independent in many species, therefore sex determination region/chromosome was different among them. Idesia polycarpa is a deciduous tree, making it an attrac- tive model system with which to study sex determination mechanism. We characterized the re-sequence data of female and male individuals from natural population. We used these data to identify the sex determination systems by statistics unique sequences in females and males. To detect the sex chromosome, we analysed the single nucleotide polymor- phisms (SNPs) by genome-wide association study (GWAS).
RESULTS The information of genome contains the secret code of various traits, so we mined the re-sequenced data of females and males. We calculated the common and unique reads in females and males, compared the unique reads between them, and found that the unique reads of females were much more than that of males. The result suggested that the sex determination system of I. polycarpa was ZW/ZZ. We then identified 30 million high quality SNPs in the population by mapping to the reference genome, calculated the p value of each SNP, drew a Manhattan plot. The result showed the highest peak on the end of 19 chromosome. We analysed the heterozygote of SNPs in this peak, and the result showed that the SNPs were heterozygotic in females, but homozygotic in males. The result confirmed that the sex determination system in I. polycarpa is ZW/ZZ. Further, a cleave amplified polymorphic sequences (CAPS) mo- lecular marker was developed according to the significant SNPs, and was successfully applied to different female and male individuals.
CONCLUSION In the Salicaceae family, there were two kinds of sex determination system, one is XX/XY (females are homogametic XX, and males are heterogametic XY), another is ZW/ZZ (females are heterogametic ZW, and males are homogametic ZZ). At present, most studies on the sex-determining regions and genes in the Salicaceae family are carried out by methods such as genome, transcriptome, and resequencing analyses. Our results showed that the sex determination system in I. polycarpawas ZW/ZZ, and the 19th chromosome is most likely associated with sex determination. And a CAPS molecular marker was developed, which is a simple and fast method for efficient identification sex at seedling stage. The research provided a useful way to the planting pattern of I. polycarpa.
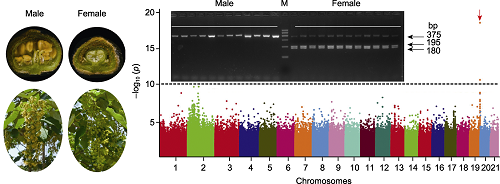
Identification of sex determination molecular marker of Idesia polycarpa. Trees of I. polycarpa are indistinguishable as to sex without examination of the flowers. A CAPS molecular marker was developed based on genome wide analysis study on a natural population.

Key words: Idesia polycarpa; sex determination; molecular marker
Yi Zuo , Hongbing Liu , Zhigang Yang , Bin Li , Haoxin Xiang , Chunzhen Zhu , Lei Wang . Identification of Sex Determination Molecular Marker Based on Genome-wide Association Study of Idesia polycarpa[J]. Chinese Bulletin of Botany, 2024 , 59(3) : 414 -421 . DOI: 10.11983/CBB24029
| [1] | 卢翔, 李效文, 郑坚, 王金旺, 夏海涛, 陈秋夏 (2010). 木本油料树种山桐子研究进展. 农业科技通讯 (5), 123-127. |
| [2] | 四川森迪科技发展股份有限公司 (2017). 一种毛叶山桐子幼苗雌雄株的鉴别方法. 中国专利, CN201710128456.X.2017-03-06. |
| [3] | 张志成, 王国礼 (1981). 一种优良的木本油料树——水冬瓜. 陕西林业科技 (5), 54-55. |
| [4] | Akagi T, Henry IM, Ohtani H, Morimoto T, Beppu K, Kataoka I, Tao R (2018). A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell 30, 780-795. |
| [5] | Akagi T, Henry IM, Tao R, Comai L (2014). A Y-chromosome- encoded small RNA acts as a sex determinant in persimmons. Science 346, 646-650. |
| [6] | Coelho SM, Mignerot L, Cock JM (2019). Origin and evo- lution of sex-determination systems in the brown algae. New Phytol 222, 1751-1756. |
| [7] | Dong N, Tang XS, Tang L (2016). Screening and analysis of sex-related ISSR molecular marker in Idesia polycarpa Maxim. var. vestita Diels. J Sichuan Univ (Nat Sci Ed) 53, 465-470. (in Chinese) |
| 董娜, 唐晓姗, 唐琳 (2016). 毛叶山桐子性别相关ISSR分子标记的筛选与分析. 四川大学学报(自然科学版) 53, 465-470 | |
| [8] | Feng C, Wang XW, Wu SS, Ning WD, Song B, Yan JB, Cheng SF (2022). HAPPE: a tool for population haplotype analysis and visualization in editable excel tables. Front Plant Sci 13, 927407. |
| [9] | Geraldes A, Hefer CA, Capron A, Kolosova N, Martinez-Nu?ez F, Soolanayakanahally RY, Stanton B, Guy RD, Mansfield SD, Douglas CJ, Cronk QCB (2015). Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol Ecol 24, 3243-3256. |
| [10] | Jeon SA, Park JL, Park SJ, Kim JH, Goh SH, Han JY, Kim SY (2021). Comparison between MGI and Illumina se- quencing platforms for whole genome sequencing. Genes Genomics 43, 713-724. |
| [11] | Lee GA, Koh HJ, Chung HK, Dixit A, Chung JW, Ma KH, Lee SY, Lee JR, Lee GS, Gwag JG, Kim TS, Park YJ (2009). Development of SNP-based CAPS and dCAPS markers in eight different genes involved in starch biosynthesis in rice. Mol Breeding 24, 93-101. |
| [12] | Li TT, Li FS, Mei LJ, Li N, Yao M, Tang L (2020). Tran- scriptome analysis of Idesia polycarpa Maxim. var vestita Diels flowers during sex differentiation. J For Res 31, 2463-2478. |
| [13] | Lu J, Chen YN, Yin TM (2021). Research progress on sex determination genes of woody plants. Chin Bull Bot 56, 90-103. (in Chinese) |
| 陆静, 陈赢男, 尹佟明 (2021). 木本植物性别决定基因研究进展. 植物学报 56, 90-103. | |
| [14] | Mei LJ, Dong N, Li FS, Li N, Yao M, Chen F, Tang L (2017). Transcriptome analysis of female and male flower buds of Idesia polycarpa Maxim. var. vestita Diels. Elect-ron J Biotechn 29, 39-46. |
| [15] | Pakull B, Groppe K, Mecucci F, Gaudet M, Sabatti M, Fladung M (2011). Genetic mapping of linkage group XIX and identification of sex-linked SSR markers in a Populus tremula × Populus tremuloides cross. Can J For Res 41, 245-253. |
| [16] | Pakull B, Groppe K, Meyer M, Markussen T, Fladung M (2009). Genetic linkage mapping in aspen (Populus tremula L. and Populus tremuloides Michx). Tree Genet Genomes 5, 505-515. |
| [17] | Paolucci I, Gaudet M, Jorge V, Beritognolo I, Terzoli S, Kuzminsky E, Muleo R, Mugnozza GS, Sabatti M (2010). Genetic linkage maps of Populus alba L. and comparative mapping analysis of sex determination across Populus species. Tree Genet Genomes 6, 863-875. |
| [18] | Peto FH (1938). Cytology of poplar species and natural hybrids. Can J Res 16c, 445-455. |
| [19] | Renner SS, Müller NA (2021). Plant sex chromosomes defy evolutionary models of expanding recombination suppression and genetic degeneration. Nat Plants 7, 392-402. |
| [20] | Renner SS, Ricklefs RE (1995). Dioecy and its correlates in the flowering plants. Am J Bot 82, 596-606. |
| [21] | Sabatti M, Gaudet M, Müller NA, Kersten B, Gaudiano C, Mugnozza GS, Fladung M, Beritognolo I (2020). Long- term study of a subdioecious Populus × canescens family reveals sex lability of females and reproduction behaviour of cosexual plants. Plant Rep 33, 1-17. |
| [22] | Sanderson BJ, Feng GQ, Hu N, Carlson CH, Smart LB, Keefover-Ring K, Yin TM, Ma T, Liu JQ, DiFazio SP, Olson MS (2021). Sex determination through X-Y heterogamety in Salix nigra. Heredity 126, 630-639. |
| [23] | Shu YJ, Li Y, Zhu ZL, Bai X, Cai H, Ji W, Guo DJ, Zhu YM (2011). SNPs discovery and CAPS marker conversion in soybean. Mol Biol Rep 38, 1841-1846. |
| [24] | Stratton M (2008). Genome resequencing and genetic variation. Nat Biotechnol 26, 65-66. |
| [25] | Walas ?, Mandryk W, Thomas PA, Tyra?a-Wierucka ?, Iszkulo G (2018) Sexual systems in gymnosperms: a re- view. Basic Appl Ecol 31, 1-9. |
| [26] | Wang SH, Li Y, Li ZQ, Chang L, Li L (2015). Identification of an SCAR marker related to female phenotype in Idesia polycarpa Maxim. Genet Mol Res 14, 2015-2022. |
| [27] | Wu ZW, Xie SX, Liu Q, Du WJ (2010). Research progress and application prospect of Idesia polycarpa. Guizhou Ag- ric Sci 38, 161-164. (in Chinese) |
| 吴志文, 谢双喜, 刘青, 杜文军 (2010). 山桐子的研究进展及应用前景. 贵州农业科学 38, 161-164. | |
| [28] | Xue LJ, Wu HT, Chen YN, Li XP, Hou J, Lu J, Wei SY, Dai XG, Olson MS, Liu JQ, Wang MX, Charlesworth D, Yin TM (2020). Two antagonistic effect genes mediate sepa- ration of sexes in a fully dioecious plant. bioRxiv. [2020- 03-15]. https://doi.org/10.1101/2020.03.15.993022 |
| [29] | Yan L (2018). Research and Development of Sex-related Molecular Markers in Idesia polycarpa. Master’s thesis. Wuhan: Yangtze University. pp. 17-19. (in Chinese) |
| 严莉 (2018). 山桐子性别相关分子标记的研究与开发. 硕士论文. 武汉: 长江大学. pp. 17-19. | |
| [30] | Yin TM, DiFazio SP, Gunter LE, Zhang XY, Sewell MM, Woolbright SA, Allan GJ, Kelleher CT, Douglas CJ, Wang MX, Tuskan GA (2008). Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res 18, 422-430. |
| [31] | Zhang X, Lü MR (2021). Component analysis, development and utilization of Idesia polycarpa. Cereal Food Ind 28(2), 53-57, 60. (in Chinese) |
| 张旋, 吕名蕊 (2021). 毛叶山桐子成分分析及开发利用. 粮食与食品工业 28(2), 53-57, 60. | |
| [32] | Zhao YH, Li XX, Chen Z, Lu HW, Liu YC, Zhang ZF, Liang CZ (2020). An overview of genome-wide association studies in plants. Chin Bull Bot 55, 715-732. (in Chinese) |
| 赵宇慧, 李秀秀, 陈倬, 鲁宏伟, 刘羽诚, 张志方, 梁承志 (2020). 生物信息学分析方法I: 全基因组关联分析概述. 植物学报 55, 715-732. | |
| [33] | Zuo Y, Liu HB, Li B, Zhao H, Li XL, Chen JT, Wang L, Zheng QB, He YQ, Zhang JS, Wang MX, Liang CZ, Wang L (2024). The Idesia polycarpa genome provides insights into its evolution and oil biosynthesis. Cell Rep 43, 113909. |
/
| 〈 |
|
〉 |