

Halo-tag Labeling Technology and It’s Application in Plant Living Cell Imaging
Received date: 2022-09-29
Accepted date: 2023-01-28
Online published: 2023-02-10
The cell is the basic structural and functional unit of living organisms, its complex internal organizational structure, interaction and process dynamics determine the life form and life process of the entire organism. The study of cell structure and function in the living state is of great significance for exploring and grasping the essence of life. With the continuous advances in science and technology, a series of new live cell labeling technologies have been innovated. In recent years, the Halo-tag labeling technology has gradually been developed into a widely used new molecular labeling technology, playing an increasingly important role in the field of living cell imaging analysis and tracking research. It has been gradually applied in plant cell imaging. In this review, we focus on the definition, classification and development process of Halo-tag technology, and introduce the reaction mechanism of the dehydrohalogenation of Halo-tag labeling technology and the types of fluorescent ligands in detail. Finally, we introduce the latest progress of this technology in the application of living cell imaging in plants is emphatically expounded from three aspects: observing the fine localization of molecules, tracking the real-time motion of molecules and detecting the interactions between molecules. Taken together, this study provides theoretical foundation and technical support for the potential application of the subsequent Halo-tag labeling technology in plants.
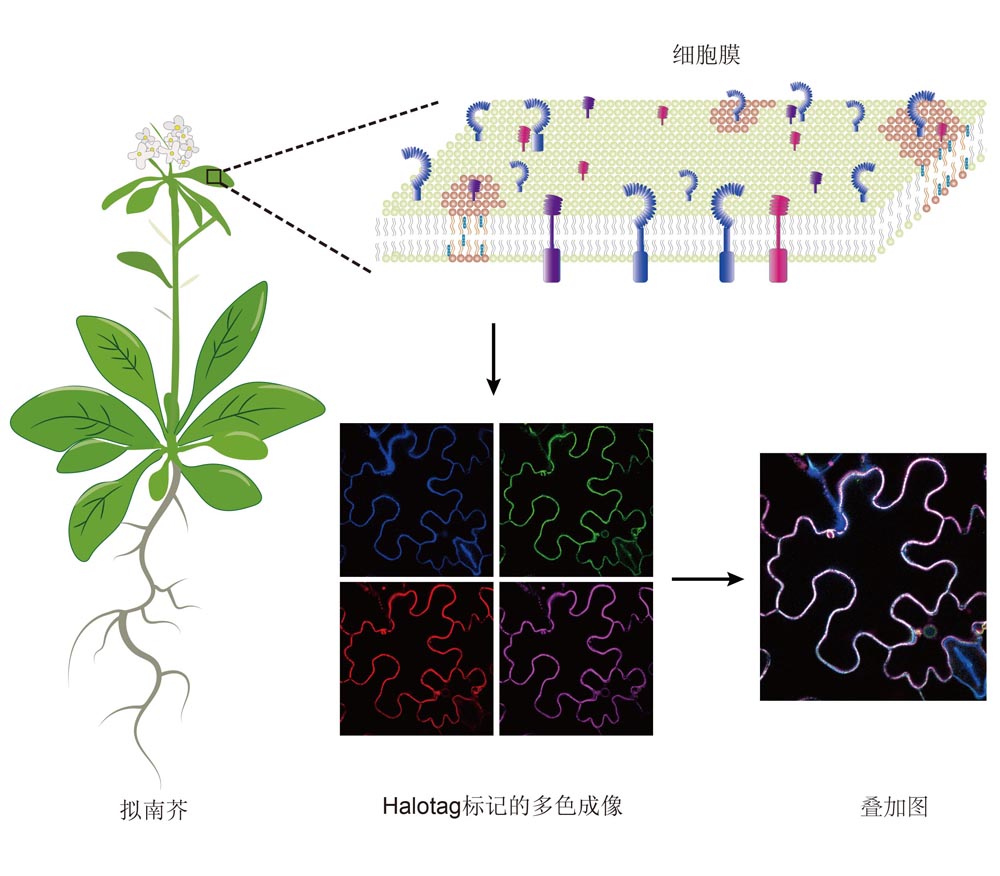
Hongping Qian , Pengyun Luo , Shuai Liu , Changwen Xu , Jinhuan Yin , Yaning Cui , Jinxing Lin . Halo-tag Labeling Technology and It’s Application in Plant Living Cell Imaging[J]. Chinese Bulletin of Botany, 2023 , 58(3) : 475 -485 . DOI: 10.11983/CBB22227
| [1] | 邝嘉怡, 李洪清, 沈文锦, 高彩吉 (2021). 基于TurboID的植物蛋白邻近标记实验方法. 植物学报 56, 584-593. |
| [2] | 李晓娟, 王钦丽, 苏月, 林金星 (2009). 量子点标记及其在植物细胞生物学中的应用. 电子显微学报 28, 495-504. |
| [3] | 钱虹萍, 陈博, 林金星, 崔亚宁 (2021). RNA聚合酶II动态调控及其成像技术的研究进展. 生物技术通报 37, 293-302. |
| [4] | 钱前, 漆小泉, 林荣呈, 杨淑华, 董爱武, 左建儒, 陈凡, 萧浪涛, 顾红雅, 陈之端, 白永飞, 王小菁, 王雷, 姜里文, 种康, 王台 (2019). 2018年中国植物科学若干领域重要研究进展. 植物学报 54, 405-440. |
| [5] | 王锋, 高婧, 王宏达 (2014). Halotag多功能标记系统及其在生物学中的应用. 电子显微学报 33, 147-155. |
| [6] | 吴凡, 沈锦波, 胡帅 (2022). 植物ESCRT复合体的功能研究进展. 植物学报 57, 697-712. |
| [7] | 夏鹏, 窦震, 姚雪彪 (2015). 超高分辨率显微技术研究进展. 生命的化学 35, 430-437. |
| [8] | 张娇, 何勤, 武泽凯, 于斌, 屈军乐, 林丹樱 (2021). 超分辨显微成像技术在活细胞成像中的应用与发展. 生物化学与生物物理进展 48, 1301-1315. |
| [9] | 张原, 张雪萍, 张月倩, 李晓娟 (2022). 单分子荧光检测技术的发展及其在植物生物学研究中的应用. 生物技术通报 38, 33-43. |
| [10] | Ai X, Fischer P, Palyha OC, Wisniewski D, Hubbard B, Akinsanya K, Strack AM, Ehrhardt AG (2012). Utilizing HaloTag technology to track the fate of PCSK9 from intracellular vs. extracellular sources. Curr Chem Genomics 6, 38-47. |
| [11] | Banaz N, M?kel? J, Uphoff S (2019). Choosing the right label for single-molecule tracking in live bacteria: side-by- side comparison of photoactivatable fluorescent protein and Halo tag dyes. J Phys D Appl Phys 52, 064002. |
| [12] | Benink HA, Urh M (2015). HaloTag technology for specific and covalent labeling of fusion proteins. In: Gautier A, Hinner MJ, eds. Site-Specific Protein Labeling: Methods and Protocols. New York: Humana. pp. 119-128. |
| [13] | Blackstock D, Chen W (2014). Halo-tag mediated self- labeling of fluorescent proteins to molecular beacons for nucleic acid detection. Chem Commun 50, 13735-13738. |
| [14] | Bruchez M Jr, Moronne M, Gin P, Weiss S, Alivisatos AP (1998). Semiconductor nanocrystals as fluorescent biological labels. Science 281, 2013-2016. |
| [15] | Cao Y, Dong HF, Pu ST, Zhang XJ (2018). Photoluminescent two-dimensional SiC quantum dots for cellular imaging and transport. Nano Res 11, 4074-4081. |
| [16] | Charubin K, Streett H, Papoutsakis ET (2020). Development of strong anaerobic fluorescent reporters for Clostridium acetobutylicum and Clostridium ljungdahlii using HaloTag and SNAP-tag proteins. Appl Environ Microbiol 86, e01271-20. |
| [17] | Chen H, Zhou Z, Li ZY, He XJ, Shen JL (2021). Highly sensitive fluorescent sensor based on coumarin organic dye for pyrophosphate ion turn-on biosensing in synovial fluid. Spectrochim Acta A Mol Biomol Spectrosc 257, 119792. |
| [18] | Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, Cisse II (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412-415. |
| [19] | Cui YN, Zhang X, Yu M, Zhu YF, Xing JJ, Lin JX (2019). Techniques for detecting protein-protein interactions in living cells: principles, limitations, and recent progress. Sci China Life Sci 62, 619-632. |
| [20] | Deprey K, Kritzer JA (2021). HaloTag forms an intramolecular disulfide. Bioconjugate Chem 32, 964-970. |
| [21] | England CG, Luo HM, Cai WB (2015). HaloTag technology: a versatile platform for biomedical applications. Bioconjugate Chem 26, 975-986. |
| [22] | Erdmann RS, Baguley SW, Richens JH, Wissner RF, Xi ZQ, Allgeyer ES, Zhong S, Thompson AD, Lowe N, Butler R, Bewersdorf J, Rothman JE, St Johnston D, Schepartz A, Toomre D (2019). Labeling strategies matter for super-resolution microscopy: a comparison between HaloTags and SNAP-tags. Cell Chem Biol 26, 584-592. |
| [23] | Feng SY, Sekine S, Pessino V, Li H, Leonetti MD, Huang B (2017). Improved split fluorescent proteins for endogenous protein labeling. Nat Commun 8, 370. |
| [24] | Fischer S, Ward TR, Liang AD (2021). Engineering a metathesis-catalyzing artificial metalloenzyme based on HaloTag. ACS Catal 11, 6343-6347. |
| [25] | Grimm JB, Brown TA, English BP, Lionnet T, Lavis LD (2017a). Synthesis of Janelia Fluor HaloTag and SNAP- tag ligands and their use in cellular imaging experiments. In: Erfle H, ed. Super-Resolution Microscopy: Methods and Protocols. New York: Humana. pp. 179-188. |
| [26] | Grimm JB, English BP, Chen JJ, Slaughter JP, Zhang ZJ, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, Lionnet T, Lavis LD (2015). A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods 12, 244-250. |
| [27] | Grimm JB, English BP, Choi H, Muthusamy AK, Mehl BP, Dong P, Brown TA, Lippincott-Schwartz J, Liu Z, Lionnet T, Lavis LD (2016). Bright photoactivatable fluorophores for single-molecule imaging. Nat Methods 13, 985-988. |
| [28] | Grimm JB, Muthusamy AK, Liang YJ, Brown TA, Lemon WC, Patel R, Lu RW, Macklin JJ, Keller PJ, Ji N, Lavis LD (2017b). A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods 14, 987-994. |
| [29] | Guo AY, Zhang YM, Wang L, Bai D, Xu YP, Wu WQ (2021). Single-molecule imaging in living plant cells: a methodological review. Int J Mol Sci 22, 5071. |
| [30] | Halstead JM, Wilbertz JH, Wippich F, Lionnet T, Ephrussi A, Chao JA (2016). TRICK: a single-molecule method for imaging the first round of translation in living cells and animals. Methods Enzymol 572, 123-157. |
| [31] | Hoelzel CA, Zhang X (2020). Visualizing and manipulating biological processes by using HaloTag and SNAP-Tag technologies. ChemBioChem 21, 1935-1946. |
| [32] | Jing YY, Zhou LL, Chen JL, Xu HJ, Sun JY, Cai MJ, Jiang JG, Gao J, Wang HD (2020). Quantitatively mapping the assembly pattern of EpCAM on cell membranes with peptide probes. Anal Chem 92, 1865-1873. |
| [33] | Kashapov RR, Bekmukhametova AM, Petrov KA, Nizameev IR, Kadirov MK, Zakharova LY (2018). Supramolecular strategy to construct quantum dot-based sensors for detection of paraoxon. Sens Actuators B Chem 273, 592-599. |
| [34] | Knight SC, Xie LQ, Deng WL, Guglielmi B, Witkowsky LB, Bosanac L, Zhang ET, El Beheiry M, Masson JB, Dahan M, Liu Z, Doudna JA, Tjian R (2015). Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science 350, 823-826. |
| [35] | Lang C, Schulze J, Mendel RR, H?nsch R (2006). HalotagTM: a new versatile reporter gene system in plant cells. J Exp Bot 57, 2985-2992. |
| [36] | Li CG, Tebo AG, Gautier A (2017a). Fluorogenic labeling strategies for biological imaging. Int J Mol Sci 18, 1473. |
| [37] | Li K, Hou JT, Yang J, Yu XQ (2017b). A tumor-specific and mitochondria-targeted fluorescent probe for real-time sensing of hypochlorite in living cells. Chem Commun (Camb) 53, 5539-5541. |
| [38] | Liu H, Dong P, Ioannou MS, Li L, Shea J, Pasolli HA, Grimm JB, Rivlin PK, Lavis LD, Koyama M, Liu Z (2018). Visualizing long-term single-molecule dynamics in vivo by stochastic protein labeling. Proc Natl Acad Sci USA 115, 343-348. |
| [39] | Liu J, Cui ZQ (2020). Fluorescent labeling of proteins of interest in live cells: beyond fluorescent proteins. Bioconjugate Chem 31, 1587-1595. |
| [40] | Liu Y, Miao K, Dunham NP, Liu HB, Fares M, Boal AK, Li XS, Zhang X (2017). The cation-π interaction enables a Halo-tag fluorogenic probe for fast no-wash live cell imaging and gel-free protein quantification. Biochemistry 56, 1585-1595. |
| [41] | Liu Z, Legant WR, Chen BC, Li L, Grimm JB, Lavis LD, Betzig E, Tjian R (2014). 3D imaging of Sox2 enhancer clusters in embryonic stem cells. eLife 3, e04236. |
| [42] | Locatelli-Hoops S, Sheen FC, Zoubak L, Gawrisch K, Yeliseev AA (2013). Application of HaloTag technology to expression and purification of cannabinoid receptor CB2. Protein Expr Purif 89, 62-72. |
| [43] | Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV (2008). HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol 3, 373-382. |
| [44] | Minner-Meinen R, Weber JN, Albrecht A, Matis R, Behnecke M, Tietge C, Frank S, Schulze J, Buschmann H, Walla PJ, Mendel RR, H?nsch R, Kaufholdt D (2021). Split-HaloTag imaging assay for sophisticated microscopy of protein-protein interactions in planta. Plant Commun 2, 100212. |
| [45] | Ove?ka M, Sojka J, Tichá M, Komis G, Basheer J, Marchetti C, ?amajová O, Kuběnová L, ?amaj J (2022). Imaging plant cells and organs with light-sheet and super-resolution microscopy. Plant Physiol 188, 683-702. |
| [46] | Pulsipher A, Griffin ME, Stone SE, Hsieh-Wilson LC (2015). Long-lived engineering of glycans to direct stem cell fate. Angew Chem Int Ed 54, 1466-1470. |
| [47] | Rasheed T, Li CL, Zhang YL, Nabeel F, Peng JX, Qi J, Gong LD, Yu CY (2018). Rhodamine-based multianalyte colorimetric probe with potentialities as on-site assay kit and in biological systems. Sens Actuators B Chem 258, 115-124. |
| [48] | Rodermund L, Coker H, Oldenkamp R, Wei GF, Bowness J, Rajkumar B, Nesterova T, Susano Pinto DM, Schermelleh L, Brockdorff N (2021). Time-resolved structured illumination microscopy reveals key principles of Xist RNA spreading. Science 372, eabe7500. |
| [49] | Shao SP, Xue BX, Sun YJ (2018). Intranucleus single- molecule imaging in living cells. Biophys J 115, 181-189. |
| [50] | Shao SP, Zhang HC, Zeng Y, Li YL, Sun CY, Sun YJ (2021). TagBiFC technique allows long-term single-molecule tracking of protein-protein interactions in living cells. Commun Biol 4, 378. |
| [51] | Shao SP, Zhang WW, Hu H, Xue BX, Qin JS, Sun CY, Sun YA, Wei WS, Sun YJ (2016). Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res 44, e86. |
| [52] | Shen WW, Ma LY, Zhang X, Li XX, Zhao YY, Jing YP, Feng Y, Tan XK, Sun F, Lin JX (2020). Three-dimensional reconstruction of Picea wilsonii Mast. pollen grains using automated electron microscopy. Sci China Life Sci 63, 171-179. |
| [53] | Simpson LM, Macartney TJ, Nardin A, Fulcher LJ, R?th S, Testa A, Maniaci C, Ciulli A, Ganley IG, Sapkota GP (2020). Inducible degradation of target proteins through a tractable affinity-directed protein missile system. Cell Chem Biol 27, 1164-1180. |
| [54] | Teng IT, Bu XN, Chung I (2019). Conjugation of fab' fragments with fluorescent dyes for single-molecule tracking on live cells. Bio Protoc 9, e3375. |
| [55] | Verschueren KHG, Seljée F, Rozeboom HJ, Kalk KH, Dijkstra BW (1993). Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature 363, 693-698. |
| [56] | Wang ZM, Detomasi TC, Chang CJ (2021). A dual-fluorophore sensor approach for ratiometric fluorescence imaging of potassium in living cells. Chem Sci 12, 1720-1729. |
| [57] | Wilhelm J, Kühn S, Tarnawski M, Gotthard G, Tünnermann J, T?nzer T, Karpenko J, Mertes N, Xue L, Uhrig U, Reinstein J, Hiblot J, Johnsson K (2021). Kinetic and structural characterization of the self-labeling protein tags HaloTag7, SNAP-tag, and CLIP-tag. Biochemistry 60, 2560-2575. |
| [58] | Yagi N, Yoshinari A, Iwatate RJ, Isoda R, Frommer WB, Nakamura M (2021). Advances in synthetic fluorescent probe labeling for live-cell imaging in plants. Plant Cell Physiol 62, 1259-1268. |
| [59] | Yan Q, Bruchez MP (2015). Advances in chemical labeling of proteins in living cells. Cell Tissue Res 360, 179-194. |
| [60] | Yazaki J, Kawashima Y, Ogawa T, Kobayashi A, Okoshi M, Watanabe T, Yoshida S, Kii I, Egami S, Amagai M, Hosoya T, Shiroguchi K, Ohara O (2020). HaloTag- based conjugation of proteins to barcoding-oligonucleotides. Nucleic Acids Res 48, e8. |
| [61] | Zhang X, Man Y, Zhuang XH, Shen JB, Zhang Y, Cui YN, Yu M, Xing JJ, Wang GC, Lian N, Hu ZJ, Ma LY, Shen WW, Yang SY, Xu HM, Bian JH, Jing YP, Li XJ, Li RL, Mao TL, Jiao YL, Sodmergen, Ren HY, Lin JX (2021). Plant multiscale networks: charting plant connectivity by multi-level analysis and imaging techniques. Sci China Life Sci 64, 1392-1422. |
| [62] | Zhang Y, Lu YQ, El Sayyed H, Bian JH, Lin JX, Li XJ (2022). Transcription factor dynamics in plants: insights and technologies for in vivo imaging. Plant Physiol 189, 23-36. |
/
| 〈 |
|
〉 |