

Research Progress and Future Perspective of Sweet Sorghum Breeding
Received date: 2022-07-15
Accepted date: 2022-11-17
Online published: 2022-11-17
Sorghum bicolor has the characteristics of salinity tolerance, high biomass and high photosynthetic performance. The stem accumulates high amounts of juice and sugar, making it an important silage crop with development potential. In-depth analysis of the molecular basis underlying the formation of S. bicolor forage traits and stress tolerance traits, as well as improvement and breeding of S. bicolor new varieties for forage are of great significance to the development of pasture-livestock industry in China. This review summarizes the research and breeding progress of S. bicolor, analyzes the existing problems, and puts forward countermeasures for S. bicolor molecular breeding in the future, aiming to promote the development of forage S. bicolor industry and ensure the food security in our country.
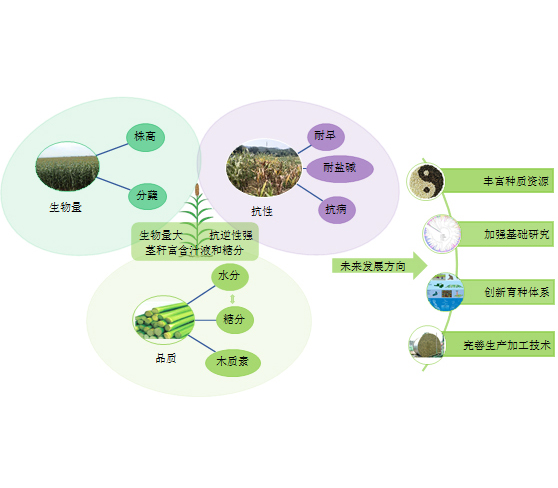
Key words: breeding; germplasm resources; pasture-livestock industry; silage; sweet sorghum
Huaiqing Hao, Ru Zhang, Cheng Lu, Hong Luo, Zhigang Li, Li Shang, Ning Wang, Zhiquan Liu, Xiaoyuan Wu, Haichun Jing . Research Progress and Future Perspective of Sweet Sorghum Breeding[J]. Chinese Bulletin of Botany, 2022 , 57(6) : 774 -784 . DOI: 10.11983/CBB22153
| [1] | 贾鹏, 王文丹, 吴天佑, 苏衍菁 (2021). 甜高粱青贮加工技术及其在反刍动物中的应用. 家畜生态学报 42(2), 86-90. |
| [2] | 景海春, 刘智全, 张丽敏, 吴小园 (2018). 饲草甜高粱分子育种与产业化. 科学通报 63, 1664-1676. |
| [3] | 景海春, 田志喜, 种康, 李家洋 (2021). 分子设计育种的科技问题及其展望概论. 中国科学: 生命科学 51, 1356-1365. |
| [4] | 黎大爵, 廖馥荪 (1992). 甜高粱及其利用. 北京: 科学出版社. pp. 144. |
| [5] | 刘杰, 黄学辉 (2021). 作物杂种优势研究现状与展望. 中国科学: 生命科学 51, 1396-1404. |
| [6] | 刘宣雨, 王青云, 刘树君, 宋松泉 (2011). 高粱遗传转化研究进展. 植物学报 46, 216-223. |
| [7] | 路登佑, 李玉蓉, 赵文峰, 冯杰 (2011). 饲用甜高粱青贮制作技术. 贵州畜牧兽医 35(2), 53-55. |
| [8] | 卢庆善 (1998). 甜高粱研究进展. 世界农业 (5), 21-23. |
| [9] | 卢庆善 (2008). 甜高粱. 北京: 中国农业科学技术出版社. pp. 113-136. |
| [10] | 卢庆善 (2011). 高粱种质资源的创新和利用. 园艺与种苗 (5), 1-4. |
| [11] | 卢庆善, 邹剑秋, 朱凯, 张志鹏, 王艳秋 (2010). 高粱种质资源的多样性和利用. 植物遗传资源学报 11, 798-801. |
| [12] | 盘道兴, 杨云, 韦继雯, 罗文, 王晓静, 钟正阳, 陈丽莎, 伏孟瑜, 李秋叶, 刘祖文, 蓬文超, 汪兴富, 黄太勇 (2018). 青贮饲料资源开发利用的研究现状. 粮食与饲料工业 (11), 52-57. |
| [13] | 闫鸿雁, 付立中, 胡国宏, 霍福德, 栾天浩, 高敬伟 (2006). 国内外甜高粱研究现状及应用前景分析. 吉林农业科学 31, 63-65. |
| [14] | 张园园 (2012). 高粱靶斑病菌致病机理及抗病基因分子标记定位. 博士论文. 沈阳: 沈阳农业大学. pp. 3. |
| [15] | 赵利铭, 刘树君, 宋松泉 (2008). 甜高粱再生体系的建立. 植物学通报 25, 465-468. |
| [16] | Adeyanju AO, Sattler SE, Rich PJ, Rivera-Burgos LA, Xu XC, Ejeta G (2021). Sorghum brown midrib19 (Bmr19) gene links lignin biosynthesis to folate metabolism. Genes (Basel). 12, 660. |
| [17] | Aguilar-Martínez JA, Poza-Carrión C, Cubas P (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458-472. |
| [18] | Bihmidine S, Julius BT, Dweikat I, Braun DM (2016). Tonoplast sugar transporters (SbTSTs) putatively control sucrose accumulation in sweet sorghum stems. Plant Signal Behav 11, e1117721. |
| [19] | Borrell AK, Mullet JE, George-Jaeggli B, van Oosterom EJ, Hammer GL, Klein PE, Jordan DR (2014). Drought adaptation of stay-green sorghum is associated with canopy development, leaf anatomy, root growth, and water uptake. J Exp Bot 65, 6251-6263. |
| [20] | Brenton ZW, Cooper EA, Myers MT, Boyles RE, Shakoor N, Zielinski KJ, Rauh BL, Bridges WC, Morris GP, Kresovich S (2016). A genomic resource for the development, improvement, and exploitation of sorghum for bioenergy. Genetics 204, 21-33. |
| [21] | Burks PS, Kaiser CM, Hawkins EM, Brown PJ (2015). Genome wide association for sugar yield in sweet sorghum. Crop Sci 55, 2138-2148. |
| [22] | Calvi?o M, Messing J (2012). Sweet sorghum as a model system for bioenergy crops. Curr Opin Biotechnol 23, 323-329. |
| [23] | Chen J, Zhu MJ, Liu RX, Zhang MJ, Lv Y, Liu YS, Xiao X, Yuan JH, Cai HW (2020). BIOMASS YIELD 1 regulates sorghum biomass and grain yield via the shikimate pathway. J Exp Bot 71, 5506-5520. |
| [24] | de Oliveira AA, Pastina MM, de Souza VF, da Costa Parrella RA, Noda RW, Simeone MLF, Schaffert RE, de Magalh?es JV, Damasceno CMB, Margarido GRA, (2018). Genomic prediction applied to high-biomass sorghum for bioenergy production. Mol Breed 38, 49. |
| [25] | Doebley J, Stec A, Gustus C (1995). Teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141, 333-346. |
| [26] | Du YL, Zhao Q, Chen LR, Yao XD, Zhang W, Zhang B, Xie FT (2020). Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol Biochem 146, 1-12. |
| [27] | Fernandes SB, Dias KOG, Ferreira DF, Brown PJ (2018). Efficiency of multi-trait, indirect, and trait-assisted genomic selection for improvement of biomass sorghum. Theor Appl Genet 131, 747-755. |
| [28] | Fujimoto M, Sazuka T, Oda Y, Kawahigashi H, Wu JZ, Takanashi H, Ohnishi T, Yoneda JI, Ishimori M, Kajiya-Kanegae H, Hibara KI, Ishizuna F, Ebine K, Ueda T, Tokunaga T, Iwata H, Matsumoto T, Kasuga S, Yonemaru JI, Tsutsumi N (2018). Transcriptional switch for programmed cell death in pith parenchyma of sorghum stems. Proc Natl Acad Sci USA 115, E8783-E8792. |
| [29] | Girma G, Nida H, Seyoum A, Mekonen M, Nega A, Lule D, Dessalegn K, Bekele A, Gebreyohannes A, Adeyanju A, Tirfessa A, Ayana G, Taddese T, Mekbib F, Belete K, Tesso T, Ejeta G, Mengiste T (2019). A large-scale genome-wide association analyses of ethiopian sorghum landrace collection reveal loci associated with important traits. Front Plant Sci 10, 691. |
| [30] | Guo W, Chen LM, Herrera-Estrella L, Cao D, Tran LSP (2020). Altering plant architecture to improve performance and resistance. Trends Plant Sci 25, 1154-1170. |
| [31] | Han YC, Lv P, Hou SL, Li SY, Ji GS, Ma X, Du RH, Liu GQ (2015). Combining next generation sequencing with bulked segregant analysis to fine map a stem moisture locus in sorghum (Sorghum bicolor L. Moench). PLoS One 10, e0127065. |
| [32] | Hao HQ, Li ZG, Leng CY, Lu C, Luo H, Liu YM, Wu XY, Liu ZQ, Shang L, Jing HC (2021). Sorghum breeding in the genomic era: opportunities and challenges. Theor Appl Genet 134, 1899-1924. |
| [33] | Hayes CM, Weers BD, Thakran M, Burow G, Xin ZG, Emendack Y, Burke JJ, Rooney WL, Mullet JE (2016). Discovery of a Dhurrin QTL in sorghum: co-localization of Dhurrin biosynthesis and a novel stay-green QTL. Crop Sci 56, 104-112. |
| [34] | Lee S, Fu FY, Liao CJ, Mewa DB, Adeyanju A, Ejeta G, Lisch D, Mengiste T (2022). Broad-spectrum fungal resistance in sorghum is conferred through the complex regulation of an immune receptor gene embedded in a natural antisense transcript. Plant Cell 34, 1641-1665. |
| [35] | Lekgari AL (2010). Genetic mapping of quantitative trait loci associated with bioenergy traits, and the assessment of genetic variability in sweet sorghum (Sorghum Bicolor (L.). Moench). Ph.D. thesis. Lincoln: University of Nebraska at Lincoln. pp. 58-77. |
| [36] | Li JQ, Tang WJ, Zhang YW, Chen KN, Wang CC, Liu YL, Zhan QW, Wang CM, Wang SB, Xie SQ, Wang LH (2018). Genome-wide association studies for five forage quality-related traits in sorghum (Sorghum bicolor L.). Front Plant Sci 9, 1146. |
| [37] | Li JQ, Wang LH, Zhan QW, Liu YL, Zhang Q, Li JF, Fan FF (2015). Mapping quantitative trait loci for five forage quality traits in a sorghum-sudangrass hybrid. Genet Mol Res 14, 13266-13273. |
| [38] | Lin ZW, Li XR, Shannon LM, Yeh CT, Wang ML, Bai GH, Peng Z, Li JR, Trick HN, Clemente TE, Doebley J, Schnable PS, Tuinstra MR, Tesso TT, White F, Yu JM (2012). Parallel domestication of the Shattering1 genes in cereals. Nat Genet 44, 720-724. |
| [39] | Liu YM, Wang ZH, Wu XY, Zhu JW, Luo H, Tian DM, Li CP, Luo JC, Zhao WM, Hao HQ, Jing HC (2021). SorGSD: updating and expanding the sorghum genome science database with new contents and tools. Biotechnol Biofuels 14, 165. |
| [40] | Luo H, Zhao WM, Wang YQ, Xia Y, Wu XY, Zhang LM, Tang BX, Zhu JW, Fang L, Du ZL, Bekele WA, Tai SS, Jordan DR, Godwin ID, Snowdon RJ, Mace ES, Luo JC, Jing HC (2016). SorGSD: a sorghum genome SNP database. Biotechnol Biofuels 9, 6. |
| [41] | Mathur S, Umakanth AV, Tonapi VA, Sharma R, Sharma MK (2017). Sweet sorghum as biofuel feedstock: recent advances and available resources. Biotechnol Biofuels 10, 146. |
| [42] | McKinley B, Rooney W, Wilkerson C, Mullet J (2016). Dynamics of biomass partitioning, stem gene expression, cell wall biosynthesis, and sucrose accumulation during development of Sorghum bicolor. Plant J 88, 662-680. |
| [43] | Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, Harriman J, Glaubitz JC, Buckler ES, Kresovich S (2013). Population genomic and genome- wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci USA 110, 453-458. |
| [44] | Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS (2003). Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302, 81-84. |
| [45] | Murray SC, Rooney WL, Hamblin MT, Mitchell SE, Kresovich S (2009). Sweet sorghum genetic diversity and association mapping for brix and height. Plant Genome 2, 48-62. |
| [46] | Murray SC, Sharma A, Rooney WL, Klein PE, Mullet JE, Mitchell SE, Kresovich S (2008). Genetic improvement of sorghum as a biofuel feedstock: I. QTL for stem sugar and grain nonstructural carbohydrates. Crop Sci 48, 2165-2179. |
| [47] | Quinby JR, Karper RE (1954). Inheritance of height in sorg-hum. Agron J 46, 211-216. |
| [48] | Ribeiro MG, de Pinho Costa KA, de Souza WF, Cruvinel WS, de Silva JT, dos Santos Júnior DR (2017). Silage quality of sorghum and Urochloa brizantha cultivars monocropped or intercropped in different planting systems. Acta Sci. Anim Sci 39, 243-250. |
| [49] | Rice B, Lipka AE (2019). Evaluation of RR-BLUP genomic selection models that incorporate peak genome-wide association study signals in maize and sorghum. Plant Genome 12, 180052. |
| [50] | Ritter KB, Jordan DR, Chapman SC, Godwin ID, Mace ES, McIntyre CL (2008). Identification of QTL for sugar- related traits in a sweet × grain sorghum (Sorghum bicolor L. Moench) recombinant inbred population. Mol Breed 22, 367-384. |
| [51] | Ritter KB, McIntyre CL, Godwin ID, Jordan DR, Chapman SC (2007). An assessment of the genetic relationship between sweet and grain sorghums, within Sorghum bicolor ssp. bicolor (L.) Moench, using AFLP markers. Euphytica 157, 161-176. |
| [52] | Salas Fernandez MG, Becraft PW, Yin YH, Lübberstedt T (2009). From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci 14, 454-461. |
| [53] | Sattler SE, Saballos A, Xin ZG, Funnell-Harris DL, Vermerris W, Pedersen JF (2014). Characterization of novel sorghum brown midrib mutants from an EMS-mutagenized population. G3 (Bethesda) 4, 2115-2124. |
| [54] | Savary S, Willocquet L, Pethybridge SJ, Esker P, McRoberts N, Nelson A (2019). The global burden of pathogens and pests on major food crops. Nat Ecol Evol 3, 430-439. |
| [55] | Shen JJ, Zhang YQ, Ge DF, Wang ZY, Song WY, Gu R, Che G, Cheng ZH, Liu RY, Zhang XL (2019). CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proc Natl Acad Sci USA 116, 17105-17114. |
| [56] | Shiringani AL, Friedt W (2011). QTL for fibre-related traits in grain × sweet sorghum as a tool for the enhancement of sorghum as a biomass crop. Theor Appl Genet 123, 999-1011. |
| [57] | Shiringani AL, Frisch M, Friedt W (2010). Genetic mapping of QTLs for sugar-related traits in a RIL population of Sorghum bicolor L. Moench. Theor Appl Genet 121, 323-336. |
| [58] | Song YS, Li SM, Sui Y, Zheng HX, Han GL, Sun X, Yang WJ, Wang HL, Zhuang KY, Kong FY, Meng QW, Sui N (2022). SbbHLH85, a bHLH member, modulates resilience to salt stress by regulating root hair growth in sorghum. Theor Appl Genet 135, 201-216. |
| [59] | Su M, Li XF, Ma XY, Peng XJ, Zhao AG, Cheng LQ, Chen SY, Liu GS (2011). Cloning two P5CS genes from bioenergy sorghum and their expression profiles under abiotic stresses and MeJA treatment. Plant Sci 181, 652-659. |
| [60] | Sui N, Yang Z, Liu ML, Wang BS (2015). Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genomics 16, 534. |
| [61] | Tao YF, Luo H, Xu JB, Cruickshank A, Zhao XR, Teng F, Hathorn A, Wu XY, Liu YM, Shatte T, Jordan D, Jing HC, Mace E (2021). Extensive variation within the pan- genome of cultivated and wild sorghum. Nat Plants 7, 766-773. |
| [62] | Tovignan TK, Adoukonou-Sagbadja H, Diatta C, Clément-Vidal A, Soutiras A, Cisse N, Luquet D (2020). Terminal drought effect on sugar partitioning and metabolism is modulated by leaf stay-green and panicle size in the stem of sweet sorghum (Sorghum bicolor L. Moench). CABI Agric Biosci 1, 4. |
| [63] | Wallace JG, Rodgers-Melnick E, Buckler ES (2018). On the road to Breeding 4.0: unraveling the good, the bad, and the boring of crop quantitative genomics. Annu Rev Genet 52, 421-444. |
| [64] | Wang B, Smith SM, Li JY (2018). Genetic regulation of shoot architecture. Annu Rev Plant Biol 69, 437-468. |
| [65] | Wang LX, Lui ACW, Lam PY, Liu GQ, Godwin ID, Lo C (2020). Transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in sorghum. Plant Biotechnol J 18, 2170-2172. |
| [66] | Wang TT, Ren ZJ, Liu ZQ, Feng X, Guo RQ, Li BG, Li LG, Jing HC (2014). SbHKT1;4, a member of the high-affinity potassium transporter gene family from Sorghum bicolor, functions to maintain optimal Na+/K+ balance under Na+ stress. J Integr Plant Biol 56, 315-332. |
| [67] | Watanabe K, Guo W, Arai K, Takanashi H, Kajiya-Kanegae H, Kobayashi M, Yano K, Tokunaga T, Fujiwara T, Tsutsumi N, Iwata H (2017). High-throughput phenotyping of sorghum plant height using an unmanned aerial vehicle and its application to genomic prediction modeling. Front Plant Sci 8, 421. |
| [68] | Wei XC, Liu LL, Lu CX, Yuan F, Han GL, Wang BS (2021). SbCASP4 improves salt exclusion by enhancing the root apoplastic barrier. Planta 254, 81. |
| [69] | Wu XY, Liu YM, Luo H, Shang L, Leng CY, Liu ZQ, Li ZG, Lu XC, Cai HW, Hao HQ, Jing HC (2022). Genomic footprints of sorghum domestication and breeding selection for multiple end uses. Mol Plant 15, 537-551. |
| [70] | Wu YY, Li XR, Xiang WW, Zhu CS, Lin ZW, Wu Y, Li JR, Pandravada S, Ridder DD, Bai GH, Wang ML, Trick HN, Bean SR, Tuinstra MR, Tesso TT, Yu JM (2012). Presence of tannins in sorghum grains is conditioned by different natural alleles of Tannin1. Proc Natl Acad Sci USA 109, 10281-10286. |
| [71] | Xie P, Shi JY, Tang SY, Chen CX, Khan A, Zhang FX, Xiong Y, Li C, He W, Wang GD, Lei FM, Wu YR, Xie Q (2019). Control of bird feeding behavior by Tannin1 through modulating the biosynthesis of polyphenols and fatty acid-derived volatiles in sorghum. Mol Plant 12, 1315-1324. |
| [72] | Xie P, Tang SY, Chen CX, Zhang HL, Yu FF, Li C, Wei HM, Sui Y, Wu CY, Diao XM, Wu YR, Xie Q (2022). Natural variation in Glume Coverage 1 causes naked grains in sorghum. Nat Commun 13, 1068. |
| [73] | Xu WW, Subudhi PK, Crasta OR, Rosenow DT, Mullet JE, Nguyen HT (2000). Molecular mapping of QTLs conferring stay-green in grain sorghum (Sorghum bicolor L. Moench). Genome 43, 461-469. |
| [74] | Xu YB, Liu XG, Fu JJ, Wang HW, Wang JK, Huang CG, Prasanna BM, Olsen MS, Wang GY, Zhang AM (2020). Enhancing genetic gain through genomic selection: from livestock to plants. Plant Commun 1, 2590-3462. |
| [75] | Yang Z, Zheng HX, Wei XC, Song J, Wang BS, Sui N (2018). Transcriptome analysis of sweet sorghum inbred lines differing in salt tolerance provides novel insights into salt exclusion by roots. Plant Soil 430, 423-439. |
| [76] | Yu H, Lin T, Meng XB, Du HL, Zhang JK, Liu GF, Chen MJ, Jing YH, Kou LQ, Li XX, Gao Q, Liang Y, Liu XD, Fan ZL, Liang YT, Cheng ZK, Chen MS, Tian ZX, Wang YH, Chu CC, Zuo JR, Wan JM, Qian Q, Han B, Zuccolo A, Wing RA, Gao CX, Liang CZ, Li JY (2021). A route to de novo domestication of wild allotetraploid rice. Cell 184, 1156-1170. |
| [77] | Yu XQ, Li XR, Guo TT, Zhu CS, Wu YY, Mitchell SE, Roozeboom KL, Wang DH, Wang ML, Pederson GA, Tesso TT, Schnable PS, Bernardo R, Yu JM (2016). Genomic prediction contributing to a promising global strategy to turbocharge gene banks. Nat Plants 2, 16150. |
| [78] | Zhang D, Tang SY, Xie P, Yang DK, Wu YR, Cheng SJ, Du K, Xin PY, Chu JF, Yu FF, Xie Q (2022). Creation of fragrant sorghum by CRISPR/Cas9. J Integr Plant Biol 64, 961-964. |
| [79] | Zhang LM, Leng CY, Luo H, Wu XY, Liu ZQ, Zhang YM, Zhang H, Xia Y, Shang L, Liu CM, Hao DY, Zhou YH, Chu CC, Cai HW, Jing HC (2018). Sweet sorghum originated through selection of Dry, a plant-specific NAC transcription factor gene. Plant Cell 30, 2286-2307. |
| [80] | Zhao J, Mantilla Perez MB, Hu JY, Salas Fernandez MG (2016). Genome-wide association study for nine plant architecture traits in sorghum. Plant Genome 9, 1-14. |
| [81] | Zheng LY, Guo XS, He B, Sun LJ, Peng Y, Dong SS, Liu TF, Jiang SY, Ramachandran S, Liu CM, Jing HC (2011). Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biol 12, R114. |
/
| 〈 |
|
〉 |