

Progress in the Research on Riboflavin Biosynthesis and Function in Plants
Received date: 2022-05-25
Accepted date: 2022-09-19
Online published: 2022-09-27
Riboflavin is the precursor of flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) that serve as an indispensable cofactor to maintain normal metabolism, which plays pivotal roles in mitochondrial electron transport chain, citric acid cycle, β-oxidation of fatty acids, branched-chain amino acid catabolism, redox homeostasis, chromatin remodeling, DNA repair, apoptosis and secondary metabolite biosynthesis. Riboflavin deficiency will cause metabolic disorders and a series of defective phenotypes, and death in the most severe cases. Among the living organisms, microorganisms and plants can de novo synthesize riboflavin, but humans and animals can only obtain it from food. At present, the regulation of riboflavin biosynthesis in microorganisms has been clearly studied, but the mechanism of riboflavin transport and metabolism in plants is still not clear. Isolating riboflavin deficient mutants is crucial for analyzing the molecular mechanisms of riboflavin biosynthesis, transport, and metabolism in plants and the effect of riboflavin on plant growth and development. Here we review first the riboflavin biosynthetic pathway and its key enzymes, and then the processes of riboflavin involved in plant growth and development in detail, and finally give prospects for plant riboflavin research.
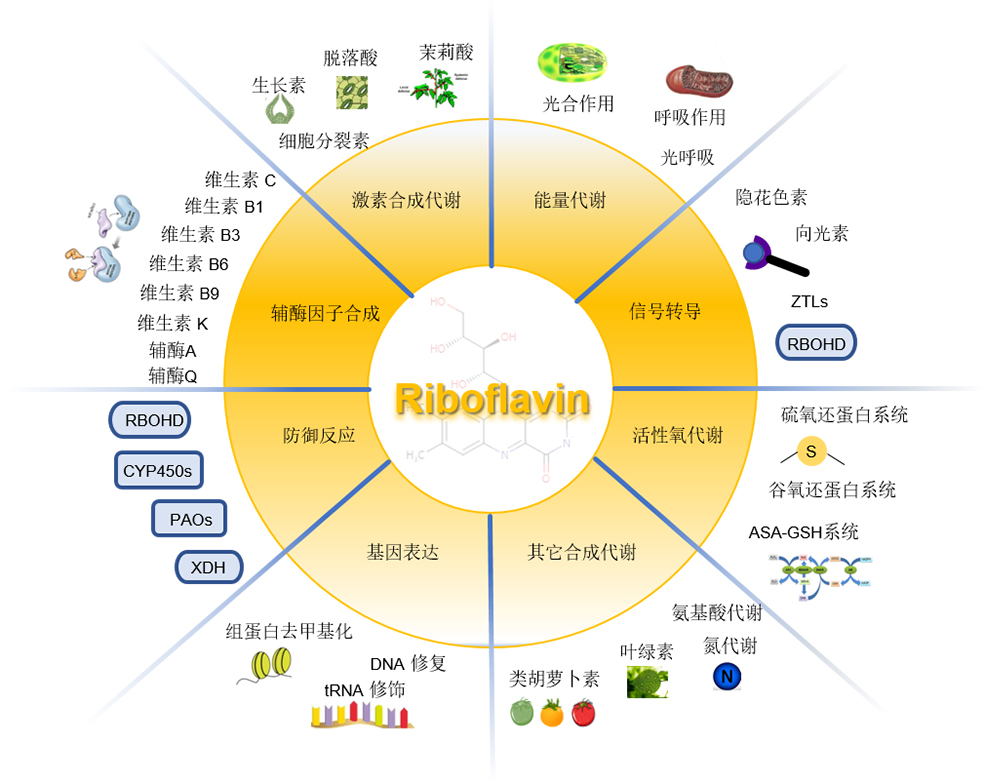
Key words: riboflavin; flavoprotein; biosynthesis; plant; transport and metabolism
Haitao Hu , Longbiao Guo . Progress in the Research on Riboflavin Biosynthesis and Function in Plants[J]. Chinese Bulletin of Botany, 2023 , 58(4) : 638 -655 . DOI: 10.11983/CBB22109
| [1] | 朱丽, 钱前 (2019). 虾青素功能米: 生物强化新思路, 优质米培育新资源. 植物学报 54, 4-8. |
| [2] | Agrawal GK, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H (2001). Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol 125, 1248-1257. |
| [3] | Averianova LA, Balabanova LA, Son OM, Podvolotskaya AB, Tekutyeva LA (2020). Production of vitamin B2 (Riboflavin) by microorganisms: an overview. Front Bioeng Biotechnol 8, 570828. |
| [4] | Balasubramaniam S, Christodoulou J, Rahman S (2019). Disorders of riboflavin metabolism. J Inherit Metab Dis 42, 608-619. |
| [5] | Barrero JM, Rodríguez PL, Quesada V, Alabadí D, Blázquez MA, Boutin JP, Marion-Poll A, Ponce MR, Micol JL (2008). The ABA1 gene and carotenoid biosynthesis are required for late skotomorphogenic growth in Arabidopsis thaliana. Plant Cell Environ 31, 227-234. |
| [6] | Blancquaert D, Storozhenko S, Loizeau K, De Steur H, De Brouwer V, Viaene J, Ravanel S, Rébeillé F, Lambert W, Van Der Straeten D (2010). Folates and folic acid: from fundamental research toward sustainable health. Crit Rev Plant Sci 29, 14-35. |
| [7] | B?ttcher C, Chapman A, Fellermeier F, Choudhary M, Scheel D, Glawischnig E (2014). The biosynthetic pathway of indole-3-carbaldehyde and indole-3-carboxylic acid derivatives in Arabidopsis. Plant Physiol 165, 841-853. |
| [8] | Brito DS, Quinhones CGS, Neri-Silva R, Heinemann B, Schertl P, Cavalcanti JHF, Eubel H, Hildebrandt T, Nunes-Nesi A, Braun HP, Araújo WL (2022). The role of the electron-transfer flavoprotein: ubiquinone oxidoreductase following carbohydrate starvation in Arabidopsis cell cultures. Plant Cell Rep 41, 431-446. |
| [9] | Bulley S, Laing W (2016). The regulation of ascorbate biosynthesis. Curr Opin Plant Biol 33, 15-22. |
| [10] | Cao X, Yang HL, Shang CQ, Ma S, Liu L, Cheng JL (2019). The roles of auxin biosynthesis YUCCA gene family in plants. Int J Mol Sci 20, 6343. |
| [11] | Chapman JM, Muhlemann JK, Gayomba SR, Muday GK (2019). RBOH-dependent ROS synthesis and ROS scavenging by plant specialized metabolites to modulate plant development and stress responses. Chem Res Toxicol 32, 370-396. |
| [12] | Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y (2020). Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol 62, 25-54. |
| [13] | Chen P, J?ger G, Zheng B (2010). Transfer RNA modifications and genes for modifying enzymes in Arabidopsis thaliana. BMC Plant Biol 10, 201. |
| [14] | Christie JM, Blackwood L, Petersen J, Sullivan S (2015). Plant flavoprotein photoreceptors. Plant Cell Physiol 56, 401-413. |
| [15] | Dai DW, Tong HY, Cheng LJ, Peng F, Zhang TT, Qi WW, Song RT (2019). Maize Dek33 encodes a pyrimidine reductase in riboflavin biosynthesis that is essential for oil- body formation and ABA biosynthesis during seed development. J Exp Bot 70, 5173-5187. |
| [16] | Delker C, Zolman BK, Miersch O, Wasternack C (2007). Jasmonate biosynthesis in Arabidopsis thaliana requires peroxisomal β-oxidation enzymes—additional proof by properties of pex6 and aim1. Phytochemistry 68, 1642-1650. |
| [17] | Dellero Y, Jossier M, Schmitz J, Maurino VG, Hodges M (2016). Photorespiratory glycolate-glyoxylate metabolism. J Exp Bot 67, 3041-3052. |
| [18] | Demarsy E, Fankhauser C (2009). Higher plants use LOV to perceive blue light. Curr Opin Plant Biol 12, 69-74. |
| [19] | Di Salvo ML, Contestabile R, Safo MK (2011). Vitamin B6 salvage enzymes: mechanism, structure and regulation. BBA-Proteins Proteom 1814, 1597-1608. |
| [20] | Ding HY, Wang BP, Han Y, Li SC (2020). The pivotal function of dehydroascorbate reductase in glutathione homeostasis in plants. J Exp Bot 71, 3405-3416. |
| [21] | Engqvist MKM, Kuhn A, Wienstroer J, Weber K, Jansen EEW, Jakobs C, Weber APM, Maurino VG (2011). Plant d-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. J Biol Chem 286, 11382-11390. |
| [22] | Fang J, Chai CL, Qian Q, Li CL, Tang JY, Sun L, Huang ZJ, Guo XL, Sun CH, Liu M, Zhang Y, Lu QT, Wang YQ, Lu CM, Han B, Chen F, Cheng ZK, Chu CC (2008). Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo- oxidation in rice. Plant J 54, 177-189. |
| [23] | Fatihi A, Latimer S, Schmollinger S, Block A, Dussault PH, Vermaas WFJ, Merchant SS, Basset GJ (2015). A dedicated type II NADPH dehydrogenase performs the penultimate step in the biosynthesis of Vitamin K1 in Synechocystis and Arabidopsis. Plant Cell 27, 1730-1741. |
| [24] | Fernie AR, Carrari F, Sweetlove LJ (2004). Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7, 254-261. |
| [25] | Grabsztunowicz M, Rantala M, Ivanauskaite A, Blomster T, Koskela MM, Vuorinen K, Tyystj?rvi E, Burow M, Overmyer K, M?h?nen AP, Mulo P (2021). Root-type ferredoxin-NADP+ oxidoreductase isoforms in Arabidopsis thaliana: expression patterns, location and stress responses. Plant Cell Environ 44, 548-558. |
| [26] | Hall M (2020). Flavoenzymes for biocatalysis. Enzymes 47, 37-62. |
| [27] | Han ML, Lv QY, Zhang J, Wang T, Zhang CX, Tan RJ, Wang YL, Zhong LY, Gao YQ, Chao ZF, Li QQ, Chen GY, Shi Z, Lin HX, Chao DY (2022). Decreasing nitrogen assimilation under drought stress by suppressing DST- mediated activation of Nitrate reductase 1.2 in rice. Mol Plant 15, 167-178. |
| [28] | Han RC, He XF, Pan XH, Shi QH, Wu ZM (2020). Enhancing xanthine dehydrogenase activity is an effective way to delay leaf senescence and increase rice yield. Rice 13, 16. |
| [29] | Hanson AD, Gregory III JF (2011). Folate biosynthesis, turnover, and transport in plants. Annu Rev Plant Biol 62, 105-125. |
| [30] | Hao JF, Pétriacq P, De Bont L, Hodges M, Gakière B (2018). Characterization of L-aspartate oxidase from Arabidopsis thaliana. Plant Sci 271, 133-142. |
| [31] | Hashida SN, Takahashi H, Uchimiya H (2009). The role of NAD biosynthesis in plant development and stress responses. Ann Bot 103, 819-824. |
| [32] | He KX, Cao XF, Deng X (2021). Histone methylation in epigenetic regulation and temperature responses. Curr Opin Plant Biol 61, 102001. |
| [33] | Hedtke B, Alawady A, Albacete A, Kobayashi K, Melzer M, Roitsch T, Masuda T, Grimm B (2012). Deficiency in riboflavin biosynthesis affects tetrapyrrole biosynthesis in etiolated Arabidopsis tissue. Plant Mol Biol 78, 77-93. |
| [34] | Herrero S, González E, Gillikin JW, Vél?z H, Daub ME (2011). Identification and characterization of a pyridoxal reductase involved in the vitamin B6 salvage pathway in Arabidopsis. Plant Mol Biol 76, 157-169. |
| [35] | Higuchi-Takeuchi M, Ichikawa T, Kondou Y, Matsui K, Hasegawa Y, Kawashima M, Sonoike K, Mori M, Hirochika H, Matsui M (2011). Functional analysis of two isoforms of leaf-type ferredoxin-NADP+-oxidoreductase in rice using the heterologous expression system of Arabidopsis. Plant Physiol 157, 96-108. |
| [36] | Hino S, Sakamoto A, Nagaoka K, Anan K, Wang YQ, Mimasu S, Umehara T, Yokoyama S, Kosai KI, Nakao M (2012). FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat Commun 3, 758. |
| [37] | Hu HT, Ren DY, Hu J, Jiang HZ, Chen P, Zeng DL, Qian Q, Guo LB (2021). WHITE AND LESION-MIMIC LEAF1, encoding a lumazine synthase, affects reactive oxygen species balance and chloroplast development in rice. Plant J 108, 1690-1703. |
| [38] | Huang SB, Millar AH (2013). Succinate dehydrogenase: the complex roles of a simple enzyme. Curr Opin Plant Biol 16, 344-349. |
| [39] | Hwang OJ, Back K (2021). Suppression of rice cryptochrome 1b decreases both melatonin and expression of brassinosteroid biosynthetic genes resulting in salt tolerance. Molecules 26, 1075. |
| [40] | Ishizaki K, Schauer N, Larson TR, Graham IA, Fernie AR, Leaver CJ (2006). The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J 47, 751-760. |
| [41] | Jiang DH, Yang WN, He YH, Amasino RM (2007). Arabidopsis relatives of the human lysine-specific demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19, 2975-2987. |
| [42] | Jones RJ, Schreiber BMN (1997). Role and function of cytokinin oxidase in plants. Plant Growth Regul 23, 123-134. |
| [43] | Joosten V, Van Berkel WJH (2007). Flavoenzymes. Curr O- pin Chem Biol 11, 195-202. |
| [44] | Jurca M, Sj?lander J, Ibá?ez C, Matrosova A, Johansson M, Kozarewa I, Takata N, Bakó L, Webb AAR, Israelsson-Nordstr?m M, Eriksson ME (2022). ZEITLUPE promotes ABA-induced stomatal closure in Arabidopsis and Populus. Front Plant Sci 13, 829121. |
| [45] | Kampire MG, Sanglou RK, Wang HM, Kazeem BB, Wu JL, Zhang XB (2021). A novel allele encoding 7-hydroxymethyl chlorophyll a reductase confers bacterial blight resistance in rice. Int J Mol Sci 22, 7585. |
| [46] | Kang ZH, Qin T, Zhao ZP (2019). Thioredoxins and thioredoxin reductase in chloroplasts: a review. Gene 706, 32-42. |
| [47] | Kong WW, Li J, Yu QY, Cang W, Xu R, Wang Y, Ji W (2016). Two novel flavin-containing monooxygenases involved in biosynthesis of aliphatic glucosinolates. Front Plant Sci 7, 1292. |
| [48] | Konishi N, Ishiyama K, Matsuoka K, Maru I, Hayakawa T, Yamaya T, Kojima S (2014). NADH-dependent glutamate synthase plays a crucial role in assimilating ammonium in the Arabidopsis root. Physiol Plant 152, 138-151. |
| [49] | Kramer M, Rodriguez-Heredia M, Saccon F, Mosebach L, Twachtmann M, Krieger-Liszkay A, Duffy C, Knell RJ, Finazzi G, Hanke GT (2021). Regulation of photosynthetic electron flow on dark to light transition by ferredoxin:NADP(H) oxidoreductase interactions. eLife 10, e56-088. |
| [50] | Kumar R, Wallis JG, Skidmore C, Browse J (2006). A mutation in Arabidopsis cytochrome b5 reductase identified by high-throughput screening differentially affects hydroxylation and desaturation. Plant J 48, 920-932. |
| [51] | Kupke T, Hernández-Acosta P, Steinbacher S, Culiá?ez- Macià FA (2001). Arabidopsis thaliana flavoprotein AtHAL3a catalyzes the decarboxylation of 4'-phosphopantothenoylcysteine to 4'-phosphopantetheine, a key step in coenzyme A biosynthesis. J Biol Chem 276, 19190-19196. |
| [52] | Lan F, Nottke AC, Shi Y (2008). Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol 20, 316-325. |
| [53] | Lermontova I, Grimm B (2006). Reduced activity of plastid protoporphyrinogen oxidase causes attenuated photodynamic damage during high-light compared to low-light exposure. Plant J 48, 499-510. |
| [54] | Li C, Wang X, Zhang LY, Zhang CY, Yu CS, Zhao T, Liu B, Li HY, Liu J (2022). OsBIC1 directly interacts with OsCRYs to regulate leaf sheath length through mediating GA-responsive pathway. Int J Mol Sci 23, 287. |
| [55] | Li SY, Du L, Bernhardt R (2020). Redox partners: function modulators of bacterial P450 enzymes. Trends Microbiol 28, 445-454. |
| [56] | Liao GL, Chen L, He YQ, Li XS, Lv ZX, Yi SY, Zhong M, Huang CH, Jia DF, Qu XY, Xu XB (2021). Three metabolic pathways are responsible for the accumulation and maintenance of high AsA content in kiwifruit (Actinidia eriantha). BMC Genom 22, 13. |
| [57] | Liscombe DK, Kamiyoshihara Y, Ghironzi J, Kempthorne CJ, Hooton K, Bulot B, Kanellis V, McNulty J, Lam NB, Nadeau LF, Pautler M, Tieman DM, Klee HJ, Goulet C (2022). A flavin-dependent monooxygenase produces nitrogenous tomato aroma volatiles using cysteine as a nitrogen source. Proc Natl Acad Sci USA 119, e2118676119. |
| [58] | Liu S, Hu WY, Wang ZW, Chen T (2020). Production of riboflavin and related cofactors by biotechnological processes. Microb Cell Fact 19, 31. |
| [59] | Liu XJ, Hu B, Chu CC (2022). Nitrogen assimilation in plants: current status and future prospects. J Genet Genomics 49, 394-404. |
| [60] | Liu YH, Yu L, Tong JH, Ding JH, Wang RZ, Lu YS, Xiao LT (2013). Tiller number is altered in the ascorbic acid- deficient rice suppressed for L-galactono-1,4-lactone dehydrogenase. J Plant Physiol 170, 389-396. |
| [61] | Maoka T (2020). Carotenoids as natural functional pigments. J Nat Med 74, 1-16. |
| [62] | Marty L, Bausewein D, Müller C, Bangash SAK, Moseler A, Schwarzl?nder M, Müller-Schüssele SJ, Zechmann B, Riondet C, Balk J, Wirtz M, Hell R, Reichheld JP, Meyer AJ (2019). Arabidopsis glutathione reductase 2 is indispensable in plastids, while mitochondrial glutathione is safeguarded by additional reduction and transport systems. New Phytol 224, 1569-1584. |
| [63] | Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot 105, 1141-1157. |
| [64] | Meguro M, Ito H, Takabayashi A, Tanaka R, Tanaka A (2011). Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 23, 3442-3453. |
| [65] | Meyer Y, Belin C, Delorme-Hinoux V, Reichheld JP, Riondet C (2012). Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid Redox Signal 17, 1124-1160. |
| [66] | Miyazaki Y, Takase T, Kiyosue T (2015). ZEITLUPE positively regulates hypocotyl elongation at warm temperature under light in Arabidopsis thaliana. Plant Signal Behav 10, e998540. |
| [67] | Mizutani M, Sato F (2011). Unusual P450 reactions in plant secondary metabolism. Arch Biochem Biophys 507, 194-203. |
| [68] | M?glich A, Yang XJ, Ayers RA, Moffat K (2010). Structure and function of plant photoreceptors. Annu Rev Plant Biol 61, 21-47. |
| [69] | Mosegaard S, Dipace G, Bross P, Carlsen J, Gregersen N, Olsen RKJ (2020). Riboflavin deficiency-implications for general human health and inborn errors of metabolism. Int J Mol Sci 21, 3847. |
| [70] | Mulo P (2011). Chloroplast-targeted ferredoxin-NADP+ oxidoreductase (FNR): structure, function and location. BBA- Bioenergetics 1807, 927-934. |
| [71] | Nisar N, Li L, Lu S, Khin NC, Pogson B (2015). Carotenoid metabolism in plants. Mol Plant 8, 68-82. |
| [72] | Niu GQ, Zhao S, Wang L, Dong W, Liu L, He YK (2017). Structure of the Arabidopsis thaliana NADPH-cytochrome P450 reductase 2 (ATR2) provides insight into its function. FEBS J 284, 754-765. |
| [73] | Noh SW, Seo RR, Park HJ, Jung HW (2021). Two Arabidopsis homologs of human lysine-specific demethylase function in epigenetic regulation of plant defense responses. Front Plant Sci 12, 688003. |
| [74] | Oh YJ, Kim H, Seo SH, Hwang BG, Chang YS, Lee J, Lee DW, Sohn EJ, Lee SJ, Lee Y, Hwang I (2016). Cytochrome b5 reductase 1 triggers serial reactions that lead to iron uptake in plants. Mol Plant 9, 501-513. |
| [75] | Pandian BA, Sathishraj R, Djanaguiraman M, Prasad PVV, Jugulam M (2020). Role of cytochrome P450 enzymes in plant stress response. Antioxidants (Basel) 9, 454. |
| [76] | Patel MS, Nemeria NS, Furey W, Jordan F (2014). The pyruvate dehydrogenase complexes: structure-based func- tion and regulation. J Biol Chem 289, 16615-16623. |
| [77] | Pedersen L, Henriksen A (2005). Acyl-CoA oxidase 1 from Arabidopsis thaliana. Structure of a key enzyme in plant lipid metabolism. J Mol Biol 345, 487-500. |
| [78] | Piao WL, Han SH, Sakuraba Y, Paek NC (2017). Rice 7-hydroxymethyl chlorophyll a reductase is involved in the promotion of chlorophyll degradation and modulates cell death signaling. Mol Cells 40, 773-786. |
| [79] | Plantone D, Pardini M, Rinaldi G (2021). Riboflavin in neurological diseases: a narrative review. Clin Drug Investig 41, 513-527. |
| [80] | Rasmusson AG, Geisler DA, M?ller IM (2008). The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8, 47-60. |
| [81] | Rébeillé F, Ravanel S, Jabrin S, Douce R, Storozhenko S, Van Der Straeten DVD (2006). Folates in plants: biosynthesis, distribution, and enhancement. Physiol Plant 126, 330-342. |
| [82] | Rong CY, Liu YX, Chang ZY, Liu ZY, Ding YF, Ding CQ (2022). Cytokinin oxidase/dehydrogenase family genes exhibit functional divergence and overlap in rice growth and development, especially in control of tillering. J Exp Bot 73, 3552-3568. |
| [83] | Ruan JJ, Zhou YX, Zhou ML, Yan J, Khurshid M, Weng WF, Cheng JP, Zhang KX (2019). Jasmonic acid signaling pathway in plants. Int J Mol Sci 20, 2479. |
| [84] | Sa N, Rawat R, Thornburg C, Walker KD, Roje S (2016). Identification and characterization of the missing phosphatase on the riboflavin biosynthesis pathway in Arabidopsis thaliana. Plant J 88, 705-716. |
| [85] | Sagi M, Fluhr R (2006). Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol 141, 336-340. |
| [86] | Schertl P, Cabassa C, Saadallah K, Bordenave M, Savouré A, Braun HP (2014). Biochemical characterization of proline dehydrogenase in Arabidopsis mitochondria. FEBS J 281, 2794-2804. |
| [87] | Schilmiller AL, Koo AJK, Howe GA (2007). Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiol 143, 812-824. |
| [88] | Schlaich NL (2007). Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci 12, 412-418. |
| [89] | Schmülling T, Werner T, Riefler M, Krupková E, Manns IBY (2003). Structure and function of cytokinin oxidase/ dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res 116, 241-252. |
| [90] | Schwechheimer SK, Park EY, Revuelta JL, Becker J, Wittmann C (2016). Biotechnology of riboflavin. Appl Microbiol Biotechnol 100, 2107-2119. |
| [91] | Seo M, Koiwai H, Akaba S, Komano T, Oritani T, Kamiya Y, Koshiba T (2000a). Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J 23, 481-488. |
| [92] | Seo M, Peeters AJM, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JAD, Koornneef M, Kamiya Y, Koshiba T (2000b). The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97, 12908-12913. |
| [93] | Shen SQ, Peng M, Fang H, Wang ZX, Zhou S, Jing XY, Zhang M, Yang CK, Guo H, Li YF, Lei L, Shi YH, Sun YY, Liu XQ, Xu CP, Tohge T, Yuan M, Fernie AR, Ning YS, Wang GL, Luo J (2021). An Oryza-specific hydroxycinnamoyl tyramine gene cluster contributes to enhanced disease resistance. Sci Bull 66, 2369-2380. |
| [94] | Shibaya T, Hori K, Ogiso-Tanaka E, Yamanouchi U, Shu K, Kitazawa N, Shomura A, Ando T, Ebana K, Wu JZ, Yamazaki T, Yano M (2016). Hd18, encoding histone acetylase related to Arabidopsis FLOWERING LOCUS D, is involved in the control of flowering time in rice. Plant Cell Physiol 57, 1828-1838. |
| [95] | Smith EN, Schwarzl?nder M, Ratcliffe RG, Kruger NJ (2021). Shining a light on NAD- and NADP-based metabolism in plants. Trends Plant Sci 26, 1072-1086. |
| [96] | Sundin L, Vanholme R, Geerinck J, Goeminne G, H?fer R, Kim H, Ralph J, Boerjan W (2014). Mutation of the inducible ARABIDOPSIS THALIANA CYTOCHROME P450 REDUCTASE 2 alters lignin composition and improves saccharification. Plant Physiol 166, 1956-1971. |
| [97] | Suwannasom N, Kao I, Pru? A, Georgieva R, B?umler H (2020). Riboflavin: the health benefits of a forgotten natural vitamin. Int J Mol Sci 21, 950. |
| [98] | Tanaka R, Tanaka A (2007). Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58, 321-346. |
| [99] | Tani T, Sobajima H, Okada K, Chujo T, Arimura SI, Tsutsumi N, Nishimura M, Seto H, Nojiri H, Yamane H (2008). Identification of the OsOPR7 gene encoding 12- oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 227, 517-526. |
| [100] | Thodberg S, Neilson EHJ (2020). The “Green” FMOs: diversity, functionality and application of plant flavoproteins. Catalysts 10, 329. |
| [101] | Tian QZ, Wang G, Ma XX, Shen QW, Ding ML, Yang XY, Luo XL, Li RR, Wang ZH, Wang XY, Fu ZY, Yang QH, Tang JH, Wang GF (2022). Riboflavin integrates cellular energetics and cell cycle to regulate maize seed development. Plant Biotechnol J 20, 1487-1501. |
| [102] | Tian YS, Xu J, Wang B, Fu XY, Gao JJ, Han HJ, Li ZJ, Wang LJ, Zhang FJ, Zhang WH, Deng YD, Wang Y, Peng RH, Yao QH (2021). Riboflavin fortification of rice endosperm by metabolic engineering. Plant Biotechnol J 19, 1483-1485. |
| [103] | Tu B, Zhang T, Wang SG, Zhou L, Zheng L, Zhang C, Li XZ, Zhang XY, Yin JJ, Zhu XB, Yuan H, Li T, Chen WL, Qin P, Ma BT, Wang YP, Li SG (2022). Loss of Gn1a/ OsCKX2 confers heavy-panicle rice with excellent lodging resistance. J Integr Plant Biol 64, 23-38. |
| [104] | Vercellino I, Sazanov LA (2022). The assembly, regulation and function of the mitochondrial respiratory chain. Nat Rev Mol Cell Biol 23, 141-161. |
| [105] | Vorbach C, Harrison R, Capecchi MR (2003). Xanthine oxidoreductase is central to the evolution and function of the innate immune system. Trends Immunol 24, 512-517. |
| [106] | Wang Q, Lin CT (2020). Mechanisms of cryptochrome- mediated photoresponses in plants. Annu Rev Plant Biol 71, 103-129. |
| [107] | Wang RY, He F, Ning YS, Wang GL (2020). Fine-tuning of RBOH-mediated ROS signaling in plant immunity. Trends Plant Sci 25, 1060-1062. |
| [108] | Wang X, Jiang BC, Gu LF, Chen YD, Mora M, Zhu MLM, Noory E, Wang Q, Lin CT (2021). A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat Plants 7, 1397-1408. |
| [109] | Wayne LL, Wallis JG, Kumar R, Markham JE, Browse J (2013). Cytochrome b5 reductase encoded by CBR1 is essential for a functional male gametophyte in Arabidopsis. Plant Cell 25, 3052-3066. |
| [110] | Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15, 2532-2550. |
| [111] | Wheeler GL, Jones MA, Smirnoff N (1998). The biosynthetic pathway of vitamin C in higher plants. Nature 393, 365-369. |
| [112] | Wu J, Kamanga BM, Zhang WY, Xu YH, Xu L (2022). Research progress of aldehyde oxidases in plants. PeerJ 10, e13119. |
| [113] | Wu ZY, Ren H, Xiong WD, Roje S, Liu YC, Su KL, Fu CX (2018). Methylenetetrahydrofolate reductase modulates methyl metabolism and lignin monomer methylation in maize. J Exp Bot 69, 3963-3973. |
| [114] | Xiao S, Dai LY, Liu FQ, Wang ZL, Peng W, Xie DX (2004). COS1: an Arabidopsis coronatine insensitive 1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. Plant Cell 16, 1132-1142. |
| [115] | Xu JJ, Zhang XF, Jiang Y, Fan H, Li JX, Li CY, Zhao Q, Yang L, Hu YH, Martin C, Chen XY (2021). A unique flavoenzyme operates in ubiquinone biosynthesis in photosynthesis-related eukaryotes. Sci Adv 7, eabl3594. |
| [116] | Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007). Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143, 1362-1371. |
| [117] | Yamauchi T, Yoshioka M, Fukazawa A, Mori H, Nishizawa NK, Tsutsumi N, Yoshioka H, Nakazono M (2017). An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen- deficient conditions. Plant Cell 29, 775-790. |
| [118] | Yamaya T, Kusano M (2014). Evidence supporting distinct functions of three cytosolic glutamine synthetases and two NADH-glutamate synthases in rice. J Exp Bot 65, 5519-5525. |
| [119] | Yang H, Li YF, Cao YW, Shi WQ, Xie E, Mu N, Du GJ, Shen Y, Tang D, Cheng ZK (2022). Nitrogen nutrition contributes to plant fertility by affecting meiosis initiation. Nat Commun 13, 485. |
| [120] | Yang WN, Jiang DH, Jiang JF, He YH (2010). A plant- specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J 62, 663-673. |
| [121] | Yokochi Y, Fukushi Y, Wakabayashi KI, Yoshida K, Hisabori T (2021). Oxidative regulation of chloroplast enzymes by thioredoxin and thioredoxin-like proteins in Arabidopsis thaliana. Proc Natl Acad Sci USA 118, e2114952118. |
| [122] | Yoshimoto N, Onuma M, Mizuno S, Sugino Y, Nakabayashi R, Imai S, Tsuneyoshi T, Sumi SI, Saito K (2015). Identification of a flavin-containing S-oxygenating monooxygenase involved in alliin biosynthesis in garlic. Plant J 83, 941-951. |
| [123] | Yu Z, Jia DY, Liu TB (2019). Polyamine oxidases play various roles in plant development and abiotic stress tolerance. Plants(Basel) 8, 184. |
| [124] | Zeng DD, Qin R, Li M, Alamin M, Jin XL, Liu Y, Shi CH (2017). The ferredoxin-dependent glutamate synthase (OsFd-GOGAT) participates in leaf senescence and the nitrogen remobilization in rice. Mol Genet Genom 292, 385-395. |
| [125] | Zhang F, Tang WJ, Hedtke B, Zhong LL, Liu L, Peng LW, Lu CM, Grimm B, Lin RC (2014). Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc Natl Acad Sci USA 111, 2023-2028. |
| [126] | Zhang JR, Ge YY, Liu PH, Wu DT, Liu HY, Li HB, Corke H, Gan RY (2022). Biotechnological strategies of riboflavin biosynthesis in microbes. Engineering 12, 115-127. |
| [127] | Zhao D, Wang H, Li ZY, Han SN, Han C, Liu AX (2022). LC_Glucose-inhibited division protein is required for motility, biofilm formation, and stress response in Lysobacter capsici X2-3. Front Microbiol 13, 840792. |
| [128] | Zhao YD, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306-309. |
/
| 〈 |
|
〉 |