

Advances in the Regulation of Rice (Oryza sativa) Grain Shape by Auxin Metabolism, Transport and Signal Transduction
Received date: 2021-12-24
Accepted date: 2022-03-18
Online published: 2022-03-18
Rice (Oryza sativa) is a major food crop in the world. The optimization and utilization of the major yield-determining factors are important for increasing yield potential. Among these factors, seed weight is one of the most important factors determining rice production. The heritability of rice grain weight is stable, which is largely unaffected by environmental factors. Grain weight depends on grain shape, which is determined by grain length, grain width, and grain thickness, and the degree of grain filling. The growth of rice glumes and seed endosperm determines the grain shape and weight. The proliferation and expansion of glume cells affect grain development, and endosperm occupies most of the volume of mature seeds. Auxin is an important plant hormone that affects rice yield, which regulates the development of glume and endosperm after fertilization. The spatial-temporal distribution of active auxin is dynamically modulated by auxin metabolism, auxin transport and signal transduction, all of which maintain auxin at the optimal level for seed development. Here we reviewed the research progress of auxin pathways regulating rice grain shape from three aspects, auxin metabolism, auxin transport and auxin signal transduction, to provide clues for exploring the auxin regulation mechanism of grain shape and improve yield in rice.
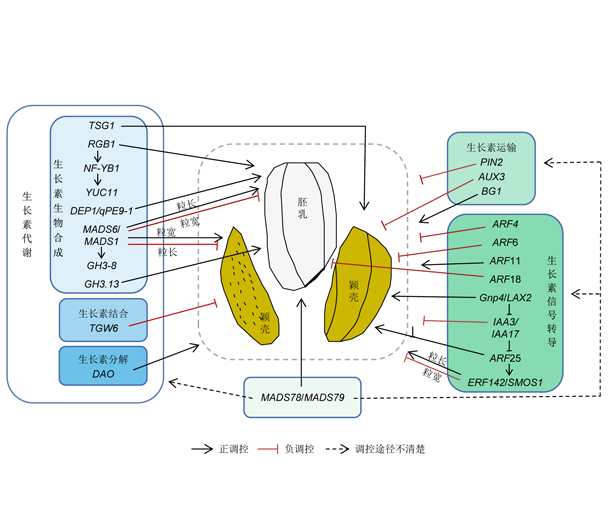
Key words: rice; auxin metabolism; auxin transport; auxin signal transduction; grain shape
Lixia Jia, Yanhua Qi . Advances in the Regulation of Rice (Oryza sativa) Grain Shape by Auxin Metabolism, Transport and Signal Transduction[J]. Chinese Bulletin of Botany, 2022 , 57(3) : 263 -275 . DOI: 10.11983/CBB21227
| [1] | 贺祯媚, 李东明, 齐艳华 (2019). 植物ABCB亚家族生物学功能研究进展. 植物学报 54, 688-698. |
| [2] | 蒋德安, 朱诚, 杨玲 (2011). 植物生理学(第2版). 北京: 高等教育出版社. pp. 168-172. |
| [3] | 林雨晴, 齐艳华 (2021). 生长素输出载体PIN家族研究进展. 植物学报 56, 151-165. |
| [4] | Abu-Zaitoon YM, Bennett K, Normanly J, Nonhebel HM (2012). A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiol Plant 146, 487-499. |
| [5] | Armengot L, Marquès-Bueno MM, Jaillais Y (2016). Regulation of polar auxin transport by protein and lipid kinases. J Exp Bot 67, 4015-4037. |
| [6] | Aya K, Hobo T, Sato-Izawa K, Ueguchi-Tanaka M, Kitano H, Matsuoka M (2014). A novel AP2-type transcription factor, SMALL ORGAN SIZE 1, controls organ size downstream of an auxin signaling pathway. Plant Cell Physiol 55, 897-912. |
| [7] | Balzan S, Johal GS, Carraro N (2014). The role of auxin transporters in monocots development. Front Plant Sci 5, 393. |
| [8] | Basunia MA, Nonhebel HM (2019). Hormonal regulation of cereal endosperm development with a focus on rice (Oryza sativa). Funct Plant Biol 46, 493-506. |
| [9] | Brumos J, Robles LM, Yun J, Vu TC, Jackson S, Alonso JM, Stepanova AN (2018). Local auxin biosynthesis is a key regulator of plant development. Dev Cell 47, 306- 318. |
| [10] | Cao JS, Li GJ, Qu DJ, Li X, Wang YN (2020). Into the seed: auxin controls seed development and grain yield. Int J Mol Sci 21, 1662. |
| [11] | Casanova-Sáez R, Mateo-Bonmatí E, Ljung K (2021). Auxin metabolism in plants. Cold Spring Harb Perspect Biol 13, a039867. |
| [12] | Chen QG, Dai XH, De-Paoli H, Cheng YF, Takebayashi Y, Kasahara H, Kamiya Y, Zhao YD (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol 55, 1072-1079. |
| [13] | Chen YN, Fan XR, Song WJ, Zhang YL, Xu GH (2012). Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol J 10, 139-149. |
| [14] | Cheng YF, Dai XH, Zhao YD (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20, 1790-1799. |
| [15] | Dai XH, Mashiguchi K, Chen QG, Kasahara H, Kamiya Y, Ojha S, DuBois J, Ballou D, Zhao YD (2013). The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J Biol Chem 288, 1448-1457. |
| [16] | Ding XH, Cao YL, Huang LL, Zhao J, Xu CG, Li XH, Wang SP (2008). Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20, 228-240. |
| [17] | Fan CC, Xing YZ, Mao HL, Lu TT, Han B, Xu CG, Li XH, Zhang QF (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112, 1164-1171. |
| [18] | French SR, Abu-Zaitoon Y, Uddin M, Bennett K, Nonhebel HM (2014). Auxin and cell wall invertase related signaling during rice grain development. Plants (Basel) 3, 95-112. |
| [19] | Gallavotti A (2013). The role of auxin in shaping shoot architecture. J Exp Bot 64, 2593-2608. |
| [20] | Gao Y, Xu H, Shen YY, Wang JB (2013). Transcriptomic analysis of rice (Oryza sativa) endosperm using the RNA- Seq technique. Plant Mol Biol 81, 363-378. |
| [21] | Garcia D, Gerald JNF, Berger F (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17, 52-60. |
| [22] | Garcia O, Bouige P, Forestier C, Dassa E (2004). Inventory and comparative analysis of rice and Arabidopsis ATP- binding cassette (ABC) systems. J Mol Biol 343, 249- 265. |
| [23] | Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271-276. |
| [24] | Guilfoyle TJ (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27, 33-43. |
| [25] | Guo T, Chen K, Dong NQ, Ye WW, Shan JX, Lin HX (2020). Tillering and small grain 1 dominates the tryptophan aminotransferase family required for local auxin biosynthesis in rice. J Integr Plant Biol 62, 581-600. |
| [26] | Gustafson FG (1936). Inducement of fruit development by growth-promoting chemicals. Proc Natl Acad Sci USA 22, 628-636. |
| [27] | Haagen-Smit AJ, Dandliker WB, Wittwer SH, Murneek AE (1946). Isolation of 3-indoleacetic acid from immature corn kernels. Am J Bot 33, 118-120. |
| [28] | Hayashi KI, Arai K, Aoi Y, Tanaka Y, Hira H, Guo RP, Hu Y, Ge CN, Zhao YD, Kasahara H, Fukui K (2021). The main oxidative inactivation pathway of the plant hormone auxin. Nat Commun 12, 6752. |
| [29] | Hirano K, Yoshida H, Aya K, Kawamura M, Hayashi M, Hobo T, Sato-Izawa K, Kitano H, Ueguchi-Tanaka M, Matsuoka M (2017). SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING form a complex to integrate auxin and brassinosteroid signaling in rice. Mol Plant 10, 590-604. |
| [30] | Hou MM, Luo FF, Wu DX, Zhang XH, Lou MM, Shen DF, Yan M, Mao CZ, Fan XR, Xu GH, Zhang YL (2021). OsPIN9, an auxin efflux carrier, is required for the regulation of rice tiller bud outgrowth by ammonium. New Phytol 229, 935-949. |
| [31] | Hu ZJ, Lu SJ, Wang MJ, He HH, Sun L, Wang HR, Liu XH, Jiang L, Sun JL, Xin XY, Kong W, Chu CC, Xue HW, Yang JS, Luo XJ, Liu JX (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol Plant 11, 736-749. |
| [32] | Huang J, Li ZY, Zhao DZ (2016). Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci Rep 6, 29938. |
| [33] | Ikeda M, Miura K, Aya K, Kitano H, Matsuoka M (2013). Genes offering the potential for designing yield-related traits in rice. Curr Opin Plant Biol 16, 213-220. |
| [34] | Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu BI, Onishia A, Miyagawa H, Katoh E (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45, 707-711. |
| [35] | Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6, 47-59. |
| [36] | Jing HW, Yang XL, Zhang J, Liu XH, Zheng HK, Dong GJ, Nian JQ, Feng J, Xia B, Qian Q, Li JY, Zuo JR (2015). Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signaling. Nat Commun 6, 7395. |
| [37] | Kabir MR, Nonhebel HM (2021). Reinvestigation of THOUSAND-GRAIN WEIGHT 6 grain weight genes in wheat and rice indicates a role in pollen development rather than regulation of auxin content in grains. Theor Appl Genet 134, 2051-2062. |
| [38] | Kakei Y, Nakamura A, Yamamoto M, Ishida Y, Yamazaki C, Sato A, Narukawa-Nara M, Soeno K, Shimada Y (2017). Biochemical and chemical biology study of rice OsTAR1 revealed that tryptophan aminotransferase is involved in auxin biosynthesis: identification of a potent OsTAR1 inhibitor, pyruvamine2031. Plant Cell Physiol 58, 598-606. |
| [39] | Lee YS, Jeong DH, Lee DY, Yi J, Ryu CH, Kim SL, Jeong HJ, Choi SC, Jin P, Yang J, Cho LH, Choi H, An G (2010). OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J 63, 18-30. |
| [40] | Li N, Li YH (2016). Signaling pathways of seed size control in plants. Curr Opin Plant Biol 33, 23-32. |
| [41] | Li PH, Li H, Liu ZJ, Zhuang Y, Wei M, Gu YY, Liu YX, Sun XQ, Tang YY, Yue L, Lu LX, Luo DG, Huang WZ, Tu SB, Wang SH (2020). Characterization of the 'Oat-Like Rice' caused by a novel allele OsMADS1Olr reveals vital importance of OsMADS1 in regulating grain shape in Oryza sativa L. Rice 13, 73. |
| [42] | Liu LC, Tong HN, Xiao YH, Che RH, Xu F, Hu B, Liang CZ, Chu JF, Li JY, Chu CC (2015). Activation of Big Grain 1 significantly improves grain size by regulating auxin transport in rice. Proc Natl Acad Sci USA 112, 11102-11107. |
| [43] | Liu XB, Wei XJ, Sheng ZH, Jiao GA, Tang SQ, Luo J, Hu PS (2016). Polycomb protein OsFIE2 affects plant height and grain yield in rice. PLoS One 11, e0164748. |
| [44] | Ljung K (2013). Auxin metabolism and homeostasis during plant development. Development 140, 943-950. |
| [45] | Mohanta TK, Mohanta N (2014). Understanding the role of Oryza sativa OsPILS (PIN Like) genes in auxin signaling. PeerJ PrePrints 2, e586v1. |
| [46] | Mohanta TK, Mohanta N, Bae H (2015). Identification and expression analysis of PIN-Like (PILS) gene family of rice treated with auxin and cytokinin. Genes 6, 622-640. |
| [47] | Paul P, Dhatt BK, Miller M, Folsom JJ, Wang Z, Krassovskaya I, Liu K, Sandhu J, Yu HH, Zhang C, Obata T, Staswick P, Walia H (2020). MADS78 and MADS79 are essential regulators of early seed development in rice. Plant Physiol 182, 933-948. |
| [48] | Petra?s?ek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, Dhonukshe P, Skupa P, Benkova E, Perry L, Krecek P, Lee OR, Fink GR, Geisler M, Murphy AS, Luschnig C, Zazi?malova E, Friml J (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914-918. |
| [49] | Qiao JY, Jiang HZ, Lin YQ, Shang LG, Wang M, Li DM, Fu XD, Geisler M, Qi YH, Gao ZY, Qian Q (2021). A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol Plant 14, 1683-1698. |
| [50] | Sabelli PA, Larkins BA (2009). The development of endosperm in grasses. Plant Physiol 149, 14-26. |
| [51] | Sakamoto T, Matsuoka M (2008). Identifying and exploiting grain yield genes in rice. Curr Opin Plant Biol 11, 209-214. |
| [52] | Salehin M, Bagchi R, Estelle M (2015). SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27, 9-19. |
| [53] | Shen CJ, Bai YH, Wang SK, Zhang SN, Wu YR, Chen M, Jiang DA, Qi YH (2010a). Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J 277, 2954-2969. |
| [54] | Shen CJ, Wang SK, Bai YH, Wu YR, Zhang SN, Chen M, Guilfoyle TJ, Wu P, Qi YH (2010b). Functional analysis of the structural domain of ARF proteins in rice (Oryza sativa L.). J Exp Bot 61, 3971-3981. |
| [55] | Sims K, Abedi-Samakush F, Szulc N, Honti MGM, Mattsson J (2021). OsARF11 promotes growth, meristem, seed, and vein formation during rice plant development. Int J Mol Sci 22, 4089. |
| [56] | Song WJ, Sun HW, Li J, Gong XP, Huang SJ, Zhu XD, Zhang YL, Xu GH (2013). Auxin distribution is differentially affected by nitrate in roots of two rice cultivars differing in responsiveness to nitrogen. Ann Bot 112, 1383- 1393. |
| [57] | Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39, 623- 630. |
| [58] | Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616-627. |
| [59] | Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Doležal K, Schlereth A, Jürgens G, Alonso JM (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177- 191. |
| [60] | Sun SY, Wang LW, Mao HL, Shao L, Li XH, Xiao JH, Ouyang YD, Zhang QF (2018). A G-protein pathway determines grain size in rice. Nat Commun 9, 851. |
| [61] | Szemenyei H, Hannon M, Long JA (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384- 1386. |
| [62] | Tabuchi H, Zhang Y, Hattori S, Omae M, Shimizu-Sato S, Oikawa T, Qian Q, Nishimura M, Kitano H, Xie H, Fang XH, Yoshida H, Kyozuka J, Chen F, Sato Y (2011). LAX PANICLE 2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276-3287. |
| [63] | Tan X, Calderon-Villalobos LI, Sharon M, Zheng CX, Robinson CV, Estelle M, Zheng N (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640-645. |
| [64] | Tao Y, Ferrer JL, Ljung K, Pojer F, Hong FX, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, Cheng YF, Lim J, Zhao YD, Ballaré CL, Sandberg G, Noel JP, Chory J (2008). Rapid synthesis of auxin via a new tryptophan- dependent pathway is required for shade avoidance in plants. Cell 133, 164-176. |
| [65] | Uchiumi T, Okamoto T (2010). Rice fruit development is associated with an increased IAA content in pollinated ovaries. Planta 232, 579-592. |
| [66] | Wang M, Qiao JY, Yu CL, Chen H, Sun CD, Huang LZ, Li CY, Geisler M, Qian Q, Jiang DA, Qi YH (2019). The auxin influx carrier, OsAUX3, regulates rice root development and responses to aluminium stress. Plant Cell Environ 42, 1125-1138. |
| [67] | Wang SK, Wu K, Yuan QB, Liu XY, Liu ZB, Lin XY, Zeng RZ, Zhu HT, Dong GJ, Qian Q, Zhang GQ, Fu XD (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44, 950-954. |
| [68] | Wang SK, Xu YX, Li ZL, Zhang SN, Lim JM, Lee KO, Li CY, Qian Q, Jiang DA, Qi YH (2014). OsMOGS is required for N-glycan formation and auxin-mediated root development in rice (Oryza sativa L.). Plant J 78, 632-645. |
| [69] | Wang YD, Zhang T, Wang RC, Zhao YD (2018). Recent advances in auxin research in rice and their implications for crop improvement. J Exp Bot 69, 255-263. |
| [70] | Wei HB, Cui BM, Ren YL, Li JH, Liao WB, Xu NF, Peng M (2006). Research progresses on auxin response factors. J Integr Plant Biol 48, 622-627. |
| [71] | Xing YZ, Zhang QF (2010). Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61, 421-442. |
| [72] | Xu M, Zhu L, Shou HX, Wu P (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol 46, 1674-1681. |
| [73] | Xu YX, Zhang SN, Guo HP, Wang SK, Xu LG, Li CY, Qian Q, Chen F, Geisler M, Qi YH, Jiang DA (2014). OsABCB14 functions in auxin transport and iron homeostasis in rice (Oryza sativa L.). Plant J 79, 106-117. |
| [74] | Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, Zhang QF (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40, 761-767. |
| [75] | Yadav SR, Khanday I, Majhi BB, Veluthambi K, Vijayraghavan U (2011). Auxin-responsive OsMGH3, a common downstream target of OsMADS1 and OsMADS6, controls rice floret fertility. Plant Cell Physiol 52, 2123- 2135. |
| [76] | Yang JC, Zhang JH, Wang ZQ, Zhu QS (2003). Hormones in the grains in relation to sink strength and postanthesis development of spikelets in rice. Plant Growth Regul 41, 185-195. |
| [77] | Ye RG, Wu YR, Gao ZY, Chen H, Jia LX, Li DM, Li XG, Qian Q, Qi YH (2021). Primary root and root hair development regulation by OsAUX4 and its participation in the phosphate starvation response. J Integr Plant Biol 63, 1555-1567. |
| [78] | Yoshikawa T, Ito M, Sumikura T, Nakayama A, Nishimura T, Kitano H, Yamaguchi I, Koshiba T, Hibara KI, Nagato Y, Itoh JI (2014). The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J 78, 927-936. |
| [79] | Yu JP, Xiong HY, Zhu XY, Zhang HL, Li HH, Miao JL, Wang WS, Tang ZS, Zhang ZY, Yao GX, Zhang Q, Pan YH, Wang X, Rashid MAR, Li JJ, Gao YM, Li ZK, Yang WC, Fu XD, Li ZC (2017). OsLD3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol 15, 28. |
| [80] | Zeng DL, Tian ZX, Rao YC, Dong GJ, Yang YL, Huang LC, Leng YJ, Xu J, Sun C, Zhang GH, Hu J, Zhu L, Gao ZY, Hu XM, Guo LB, Xiong GS, Wang YH, Li JY, Qian Q (2017). Rational design of high-yield and superior-quality rice. Nat Plants 3, 17031. |
| [81] | Zhang DP, Zhang MY, Liang JS (2021). RGB1 regulates grain development and starch accumulation through its effect on OsYUC11-mediated auxin biosynthesis in rice endosperm cells. Front Plant Sci 12, 585174. |
| [82] | Zhang DP, Zhang MY, Zhou Y, Wang YZ, Shen JY, Chen HYX, Zhang L, Lü B, Liang GH, Liang JS (2019). The Rice G protein γ subunit DEP1/qPE9-1 positively regulates grain-filling process by increasing auxin and cytokinin content in rice grains. Rice 12, 91. |
| [83] | Zhang H, Tan GL, Yang LN, Yang JC, Zhang JH, Zhao BH (2009a). Hormones in the grains and roots in relation to post-anthesis development of inferior and superior spikelets in japonica/indica hybrid rice. Plant Physiol Biochem 47, 195-204. |
| [84] | Zhang J, Nallamilli BR, Mujahid H, Peng ZH (2010). OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J 64, 604-617. |
| [85] | Zhang Q, Li JJ, Zhang WJ, Yan SN, Wang R, Zhao JF, Li YJ, Qi ZG, Sun ZX, Zhu ZG (2012). The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J 72, 805-816. |
| [86] | Zhang QF, Wing R (2013). Genome studies and molecular genetics: understanding the functional genome based on the rice model. Curr Opin Plant Biol 16, 129-132. |
| [87] | Zhang SW, Li CH, Cao J, Zhang YC, Zhang SQ, Xia YF, Sun DY, Sun Y (2009b). Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol 151, 1889-1901. |
| [88] | Zhang T, Li RN, Xing JL, Yan L, Wang RC, Zhao YD (2018a). The YUCCA-Auxin-WOX11 module controls crown root development in rice. Front Plant Sci 9, 523. |
| [89] | Zhang ZY, Li JJ, Tang ZS, Sun XM, Zhang HL, Yu JP, Yao GX, Li GL, Guo HF, Li JL, Wu HM, Huang HG, Xu YW, Yin ZG, Qi YH, Huang RF, Yang WC, Li ZC (2018b). Gnp4/LAX2, a RAWUL protein, interferes with the OsIAA3-OsARF25 interaction to regulate grain length via the auxin signaling pathway in rice. J Exp Bot 69, 4723-4737. |
| [90] | Zhang ZY, Li JJ, Yao GX, Zhang HL, Dou HJ, Shi HL, Sun XM, Li ZC (2011). Fine mapping and cloning of the grain number per-panicle gene (Gnp4) on chromosome 4 in rice (Oryza sativa L.). Agric Sci China 10, 1825-1833. |
| [91] | Zhao ZG, Zhang YH, Liu X, Zhang X, Liu SC, Yu XW, Ren YL, Zheng XM, Zhou KN, Jiang L, Guo XP, Gai Y, Wu CY, Zhai HQ, Wang HY, Wan JM (2013). A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell 27, 113-122. |
| [92] | Zhou Y, Miao J, Gu HY, Peng XR, Leburu M, Yuan FH, Gu HW, Gao Y, Tao YJ, Zhu JY, Gong ZY, Yi CD, Gu MH, Yang ZF, Liang GH (2015). Natural variations in SLG7 regulate grain shape in rice. Genetics 201, 1591-1599. |
/
| 〈 |
|
〉 |