

Recognition, Immune Activation and Signal Regulation of Plant NLR Immune Receptor
Received date: 2021-09-14
Accepted date: 2022-02-07
Online published: 2022-02-07
A large area of membrane surface and intracellular immune receptors have been evolved in higher plants to sense various pathogen signals and prevent pathogen invasion. Among them, pattern recognition receptors on the cell surface activate basic immune response after sensing pattern molecules, while nucleotide-bounding leucine-rich repeat proteins (NLRs) activate specific immune response by sensing effector proteins secreted by pathogenic microorganisms, resulting in hypersensitivity and cell death. In this review, the latest research progress of plant immunity is mainly reviewed from the aspects of NLRs on the recognition of effector proteins, plant immune activation and downstream signal regulation.
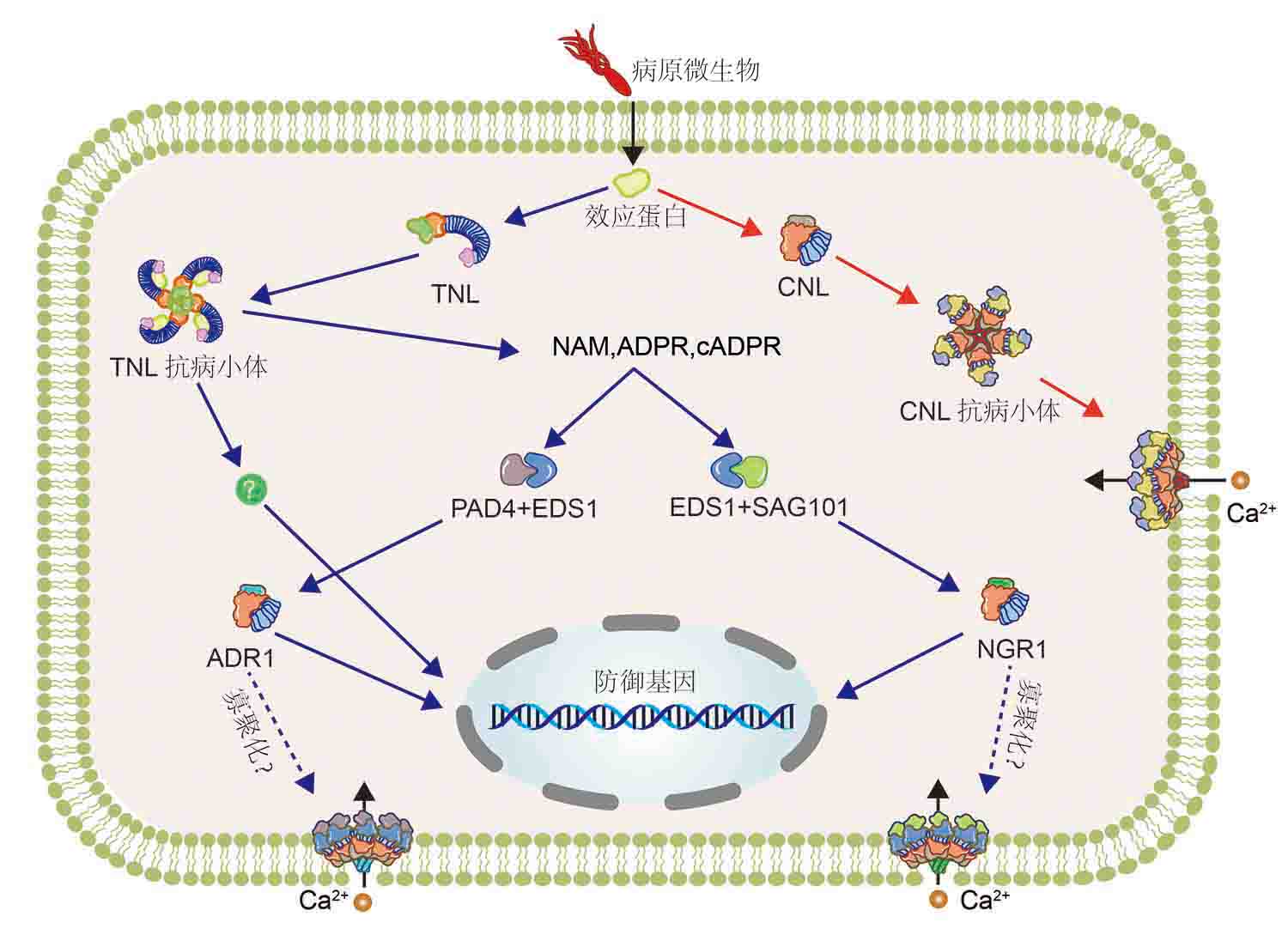
Key words: plant immunity; NLRs; receptor recognition; signal regulation
Lei Qin , Zhihong Peng , Shitou Xia . Recognition, Immune Activation and Signal Regulation of Plant NLR Immune Receptor[J]. Chinese Bulletin of Botany, 2022 , 57(1) : 12 -23 . DOI: 10.11983/CBB21159
| [1] | 王伟, 唐定中 (2021). 两类免疫受体强强联手筑牢植物免疫防线. 植物学报 56, 142-146. |
| [2] | 夏石头, 李昕 (2019). 开启防御之门: 植物抗病小体. 植物学报 54, 288-292. |
| [3] | Ade J, DeYoung BJ, Golstein C, Innes RW (2007). Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104, 2531-2536. |
| [4] | Axtell MJ, Staskawicz BJ (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369-377. |
| [5] | Bai SW, Liu J, Chang C, Zhang L, Maekawa T, Wang QY, Xiao WK, Liu YL, Chai JJ, Takken FLW, Schulze-Lefert P, Shen QH (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog 8, e1002752. |
| [6] | Bailey PC, Schudoma C, Jackson W, Baggs E, Dagdas G, Haerty W, Moscou M, Krasileva KV (2018). Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol 19, 23. |
| [7] | Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY 1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18, 1038-1051. |
| [8] | Bhandari DD, Lapin D, Kracher B, Von Born P, Bautor J, Niefind K, Parker JE (2019). An EDS1 heterodimer signaling surface enforces timely reprogramming of immunity genes in Arabidopsis. Nat Commun 10, 772. |
| [9] | Bi GZ, Su M, Li N, Liang Y, Dang S, Xu JC, Hu MJ, Wang JZ, Zou MX, Deng YN, Li QY, Huang SJ, Li JJ, Chai JJ, He KM, Chen YH, Zhou JM (2021). The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528-3541. |
| [10] | Bonardi V, Tang SJ, Stallmann A, Roberts M, Cherkis K, Dangl JL (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci USA 108, 16463-16468. |
| [11] | Castel B, Ngou PM, Cevik V, Redkar A, Kim DS, Yang Y, Ding P, Jones JDG (2019). Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol 222, 966-980. |
| [12] | Césari S, Kanzaki H, Fujiwara T, Bernoux M, Chalvon V, Kawano Y, Shimamoto K, Dodds P, Terauchi R, Kroj T (2014). The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J 33, 1941-1959. |
| [13] | Chiang YH, Coaker G (2014). Effector triggered immunity: NLR immune perception and downstream defense responses. The Arabidopsis Book 12, e0183. |
| [14] | Choi S, Prokchorchik M, Lee H, Gupta R, Lee Y, Chung EH, Cho B, Kim MS, Kim ST, Sohn KH (2021). Direct acetylation of a conserved threonine of RIN4 by the bac-terial effector HopZ5 or AvrBsT activates RPM1-dependent immunity in Arabidopsis. Mol Plant 14, 1051-1960. |
| [15] | Chung EH, Da Cunha L, Wu AJ, Gao ZY, Cherkis K, Afzal AJ, Mackey D, Dangl JL (2011). Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9, 125-136. |
| [16] | Couto D, Zipfel C (2016). Regulation of pattern recognition receptor signaling in plants. Nat Rev Immunol 16, 537-552. |
| [17] | Cui HT, Tsuda K, Parker JE (2015). Effector-triggered im-munity: from pathogen perception to robust defense. Annu Rev Plant Biol 66, 487-511. |
| [18] | Dangl JL, Jones JDG (2001). Plant pathogens and integ-rated defence responses to infection. Nature 411, 826-833. |
| [19] | Davis BK, Wen HT, Ting JPY (2011). The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol 29, 707-735. |
| [20] | De La Concepcion JC, Franceschetti M, MacLean D, Terauchi R, Kamoun S, Banfield MJ (2019). Protein engineering expands the effector recognition profile of a rice NLR immune receptor. eLife 8, e47713. |
| [21] | Ding PT, Ngou BPM, Furzer OJ, Sakai T, Shrestha RK, MacLean D, Jones JDG (2020). High-resolution expres-sion profiling of selected gene sets during plant immune activation. Plant Biotechnol J 18, 1610-1619. |
| [22] | Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CIA, Ayliffe MA, Kobe B, Ellis JG (2006). Direct protein interaction underlies gene-for-gene specificity and coevo-lution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA 103, 8888-8893. |
| [23] | Dodds PN, Rathjen JP (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11, 539-548. |
| [24] | Dong OX, Tong MMZ, Bonardi V, El Kasmi F, Woloshen V, Wünsch LK, Dangl JL, Li X (2016). TNL-mediated immunity in Arabidopsis requires complex regulation of the redundant ADR1 gene family. New Phytol 210, 960-973. |
| [25] | Duxbury Z, Wu CH, Ding PT (2021). A comparative over-view of the intracellular guardians of plants and animals: NLRs in innate immunity and beyond. Annu Rev Plant Biol 72, 155-184. |
| [26] | Essuman K, Summers DW, Sasaki Y, Mao XR, DiAntonio A, Milbrandt J (2017). The SARM1 Toll/interleukin-1 re-ceptor domain possesses intrinsic NAD+cleavage activity that promotes pathological axonal degeneration. Neuron 93, 1334-1343. |
| [27] | Feng F, Yang F, Rong W, Wu XG, Zhang J, Chen S, He CZ, Zhou JM (2012). A Xanthomonas uridine 5'-monophosphate transferase inhibits plant immune kinases. Na-ture 485, 114-118. |
| [28] | Gantner J, Ordon J, Kretschmer C, Guerois R, Stuttmann J (2019). An EDS1-SAG101 complex is essential for TNL-mediated immunity in Nicotiana benthamiana. Plant Cell 31, 2456-2474. |
| [29] | Gao ZY, Chung EH, Eitas TK, Dangl JL (2011). Plant intracellular innate immune receptor resistance to Pseudo-monas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci USA 108, 7619-7624. |
| [30] | Hu M, Qi J, Bi G, Zhou JM (2020). Bacterial effectors in-duce oligomerization of immune receptor ZAR1 in vivo. Mol Plant 13, 793-801. |
| [31] | Jacob F, Kracher B, Mine A, Seyfferth C, Blanvillain- Baufumé S, Parker JE, Tsuda K, Schulze-Lefert P, Maekawa T (2018). A dominant-interfering camta3 muta-tion compromises primary transcriptional outputs mediated by both cell surface and intracellular immune recep-tors in Arabidopsis thaliana. New Phytol 217, 1667-1680. |
| [32] | Jacob P, Kim NH, Wu F, El-Kasmi F, Chi Y, Walton WG, Furzer OJ, Lietzan AD, Sunil S, Kempthorn K, Redinbo MR, Pei ZM, Wan L, Dangl JL (2021). Plant ‘helper' im-mune receptors are Ca2+-permeable nonselective cation channels. Science 373, 420-425. |
| [33] | Jones JDG, Vance RE, Dangl JL (2016). Intracellular in-nate immune surveillance devices in plants and animals. Science 354, aaf6395. |
| [34] | Jubic LM, Saile S, Furzer OJ, El Kasmi F, Dangl JL (2019). Help wanted: helper NLRs and plant immune re-sponses. Curr Opin Plant Biol 50, 82-94. |
| [35] | Kourelis J, Van Der Hoorn RAL (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285-299. |
| [36] | Laflamme B, Dillon MM, Martel A, Almeida RND, Des-veaux D, Guttman DS (2020). The pan-genome effec-tor-triggered immunity landscape of a host-pathogen in-teraction. Science 367, 763-768. |
| [37] | Lapin D, Kovacova V, Sun XH, Dongus JA, Bhandari D, Von Born P, Bautor J, Guarneri N, Rzemieniewski J, Stuttmann J, Beyer A, Parker JE (2019). A coevolved EDS1-SAG101-NRG1 module mediates cell death sig-naling by TIR-domain immune receptors. Plant Cell 31, 2430-2455. |
| [38] | Le Roux C, Huet G, Jauneau A, Camborde L, Trémousaygue D, Kraut A, Zhou BB, Levaillant M, Adachi H, Yoshioka H, Raffaele S, Berthomé R, Couté Y, Parker JE, Deslandes L (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161, 1074-1088. |
| [39] | Lewis JD, Lee AH, Hassan JA, Wan J, Hurley B, Jhingree JR, Wang PW, Lo T, Youn JY, Guttman DS, Desveaux D (2013). The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc Natl Acad Sci USA 110, 18722-18727. |
| [40] | Liu J, Elmore JM, Lin ZJD, Coaker G (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9, 137-146. |
| [41] | Liu Y, Zeng Z, Zhang YM, Li Q, Jiang XM, Jiang Z, Tang JH, Chen DJ, Wang Q, Chen JQ, Shao ZQ (2021). An angiosperm NLR Atlas reveals that NLR gene reduction is associated with ecological specialization and signal transduction component deletion. Mol Plant 14, 2015-2031. |
| [42] | Ma S, Lapin D, Liu L, Sun Y, Song W, Zhang XX, Logemann E, Yu DL, Wang J, Jirschitzka J, Han ZF, Schulze-Lefert P, Parker JE, Chai JJ (2020). Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370, eabe3069. |
| [43] | Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379-389. |
| [44] | Martin R, Qi TC, Zhang HB, Liu FR, King M, Toth C, Nogales E, Staskawicz BJ (2020). Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 370, eabd9993. |
| [45] | Mine A, Seyfferth C, Kracher B, Berens ML, Becker D, Tsuda K (2018). The defense phytohormone signaling network enables rapid, high-amplitude transcriptional reprogramming during effector-triggered immunity. Plant Cell 30, 1199-1219. |
| [46] | Ngou BPM, Ahn HK, Ding PT, Jones JDG (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110-115. |
| [47] | Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL (2000). Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101, 353-363. |
| [48] | Ofir G, Herbst E, Baroz M, Cohen D, Millman A, Doron S, Tal N, Malheiro DBA, Malitsky S, Amitai G, Sorek R (2021). Antiviral activity of bacterial TIR domains via sig-naling molecules that trigger cell death. Nature 600, 116-120. |
| [49] | Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC (2005). NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol 15, 968-973. |
| [50] | Qi TC, Seong K, Thomazella DPT, Kim JR, Pham J, Seo E, Cho MJ, Schultink A, Staskawicz BJ (2018). NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc Natl Acad Sci USA 115, E10979-E10987. |
| [51] | Rayamajhi M, Zak DE, Chavarria-Smith J, Vance RE, Miao EA (2013). Cutting edge: mouse NAIP1 detects the type III secretion system needle protein. J Immunol 191, 3986-3989. |
| [52] | Redditt TJ, Chung EH, Karimi HZ, Rodibaugh N, Zhang YX, Trinidad JC, Kim JH, Zhou Q, Shen MZ, Dangl JL, Mackey D, Innes RW (2019). AvrRpm1 functions as an ADP-ribosyltransferase to modify NOI domain-containing proteins, including Arabidopsis and soybean RPM1-interacting protein 4. Plant Cell 31, 2664-2681. |
| [53] | Rufián JS, Rueda-Blanco J, López-Márquez D, Macho AP, Beuzón CR, Ruiz-Albert J (2021). The bacterial ef-fector HopZ1a acetylates MKK7 to suppress plant immu-nity. New Phytol 231, 1138-1156. |
| [54] | Saile SC, Jacob P, Castel B, Jubic LM, Salas-Gonzáles I, Bäcker M, Jones JDG, Dangl JL, El Kasmi F (2020). Two unequally redundant ‘helper' immune receptor fami-lies mediate Arabidopsis thaliana intracellular 'sensor' immune receptor functions. PLoS Biol 18, e3000783. |
| [55] | Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, Skle-nar J, Derbyshire P, Cevik V, Rallapalli G, Saucet SB, Wirthmueller L, Menke FLH, Sohn KH, Jones JDG (2015). A plant immune receptor detects pathogen effec-tors that target WRKY transcription factors. Cell 161, 1089-1100. |
| [56] | Saur IM, Bauer S, Kracher B, Lu XL, Franzeskakis L, Müller MC, Sabelleck B, Kümmel F, Panstruga R, Maekawa T, Schulze-Lefert P (2019). Multiple pairs of allelic MLA immune receptor-powdery mildew AVRA ef-fectors argue for a direct recognition mechanism. eLife 8, e44471. |
| [57] | Saur IML, Panstruga R, Schulze-Lefert P (2021). NOD-like receptor-mediated plant immunity: from structure to cell death. Nat Rev Immunol 21, 305-318. |
| [58] | Schultink A, Qi TC, Bally J, Staskawicz B (2019). Using forward genetics in Nicotiana benthamiana to uncover the immune signaling pathway mediating recognition of the Xanthomonas perforans effector XopJ4. New Phytol 221, 1001-1009. |
| [59] | Seto D, Koulena N, Lo T, Menna A, Guttman DS, Des-veaux D (2017). Expanded type III effector recognition by the ZAR1 NLR protein using ZED1-related kinases. Nat Plants 3, 17027. |
| [60] | Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301, 1230-1233. |
| [61] | Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, U?lker B, Somssich IE, Schulze-Lefert P (2007). Nuclear activity of MLA immune receptors links isolate- specific and basal disease-resistance responses. Science 315, 1098-1103. |
| [62] | Sun XH, Lapin D, Feehan JM, Stolze SC, Kramer K, Dongus JA, Rzemieniewski J, Blanvillain-Baufumé S, Harzen A, Bautor J, Derbyshire P, Menke FLH, Finkemeier I, Nakagami H, Jones JDG, Parker JE (2021). Pathogen effector recognition-dependent association of NRG1 with EDS1 and SAG101 in TNL receptor immunity. Nat Commun 12, 3335. |
| [63] | Tamborski J, Krasileva KV (2020). Evolution of plant NLRs: from natural history to precise modifications. Annu Rev Plant Biol 71, 355-378. |
| [64] | Thor K, Jiang SS, Michard E, George J, Scherzer S, Huang SG, Dindas J, Derbyshire P, Leitão N, DeFalco TA, Köster P, Hunter K, Kimura S, Gronnier J, Stransfeld L, Kadota Y, Bücherl CA, Charpentier M, Wrzaczek M, MacLean D, Oldroyd GED, Menke FLH, Roelfsema MRG, Hedrich R, Feijó J, Zipfel C (2020). The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 585, 569-573. |
| [65] | Tian W, Hou CC, Ren ZJ, Wang C, Zhao FG, Dahlbeck D, Hu SP, Zhang LY, Niu Q, Li LG, Staskawicz BJ, Luan S (2019). A calmodulin-gated calcium channel links patho-gen patterns to plant immunity. Nature 572, 131-135. |
| [66] | Urbach JM, Ausubel FM (2017). The NBS-LRR architect-tures of plant R-proteins and metazoan NLRs evolved in independent events. Proc Natl Acad Sci USA 114, 1063-1068. |
| [67] | Van De Weyer AL, Monteiro F, Furzer OJ, Nishimura MT, Cevik V, Witek K, Jones JDG, Dangl JL, Weigel D, Bemm F (2019). A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell 178, 1260-1272. |
| [68] | Van Der Burgh AM, Joosten MHAJ (2019). Plant immunity: thinking outside and inside the box. Trends Plant Sci 24, 587-601. |
| [69] | Van Der Hoorn RAL, Kamoun S (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009-2017. |
| [70] | Wan L, Essuman K, Anderson RG, Sasaki Y, Monteiro F, Chung EH, Nishimura EO, DiAntonio A, Milbrandt J, Dangl JL, Nishimura MT (2019). TIR domains of plant immune receptors are NAD+-cleaving enzymes that pro-mote cell death. Science 365, 799-803. |
| [71] | Wang GX, Roux B, Feng F, Guy E, Li L, Li NN, Zhang XJ, Lautier M, Jardinaud MF, Chabannes M, Arlat M, Chen S, He CZ, Noël LD, Zhou JM (2015). The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18, 285-295. |
| [72] | Wang JZ, Hu MJ, Wang J, Qi JF, Han ZF, Wang GX, Qi YJ, Wang HW, Zhou JM, Chai JJ (2019). Reconstitution and structure of a plant NLR resistosome conferring im-munity. Science 364, eaav5870. |
| [73] | Wu ZS, Li M, Dong OX, Xia ST, Liang WW, Bao YK, Wasteneys G, Li X (2019). Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol 222, 938-953. |
| [74] | Wu ZS, Tian L, Liu XR, Zhang YL, Li X (2021). TIR signal promotes interactions between lipase-like proteins and ADR1-L1 receptor and ADR1-L1 oligomerization. Plant Physiol 187, 681-686. |
| [75] | Xia BQ, Fang S, Chen XQ, Hu H, Chen PY, Wang HY, Gao ZB (2016). MLKL forms cation channels. Cell Res 26, 517-528. |
| [76] | Xia ST, Liu XR, Zhang YL (2021). Calcium channels at the center of NLR-mediated plant immunity. J Genet Genomics 48, 429-432. |
| [77] | Yin JL, Wang LQ, Jin TT, Nie Y, Liu H, Qiu YL, Yang YH, Li BW, Zhang JJ, Wang DG, Li K, Xu K, Zhi HJ (2021). A cell wall-localized NLR confers resistance to Soybean mosaic virus by recognizing viral-encoded cylindrical in-clusion protein. Mol Plant 14, 1881-1990. |
| [78] | Yoshioka K, Moeder W, Kang HG, Kachroo P, Masmoudi K, Berkowitz G, Klessig DF (2006). The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18, 747-763. |
| [79] | Yuan MH, Jiang ZY, Bi GZ, Nomura K, Liu MH, Wang YP, Cai BY, Zhou JM, He SY, Xin XF (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105-109. |
| [80] | Zhang J, Li W, Xiang TT, Liu ZX, Laluk K, Ding XJ, Zou Y, Gao MH, Zhang XJ, Chen S, Mengiste T, Zhang YL, Zhou JM (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290-301. |
| [81] | Zhao CH, Tang YH, Wang JL, Zeng YH, Sun HQ, Zheng ZC, Su R, Schneeberger K, Parker JE, Cui HT (2021). A mis-regulated cyclic nucleotide-gated channel mediates cytosolic calcium elevation and activates immunity in Arabidopsis. New Phytol 230, 1078-1094. |
| [82] | Zhou JM, Chai JJ (2008). Plant pathogenic bacterial type III effectors subdue host responses. Curr Opin Microbiol 11, 179-185. |
| [83] | Zhou JM, Zhang YL (2020). Plant immunity: danger perception and signaling. Cell 181, 978-989. |
/
| 〈 |
|
〉 |