

Research Progress of the Physiological and Molecular Mechanisms of Cadmium Accumulation in Rice
Received date: 2021-12-20
Accepted date: 2022-01-20
Online published: 2022-01-20
The wide occurrence of Cadmium (Cd)-contaminated rice in China poses significant public health risk. Breeding rice varieties with low-Cd accumulation is an effective strategy to reduce Cd accumulation in rice grain. It is necessary to understand the characteristics of Cd accumulation in rice, its physiological process and related functional genes. Here, we review the advances in physiological and molecular mechanisms of Cd uptake in roots, loading and translocation in xylem, distribution in nodes, redistribution in leaves, and accumulation in grains, which will provide theoretical reference for breeding and safe production of low-Cd rice.
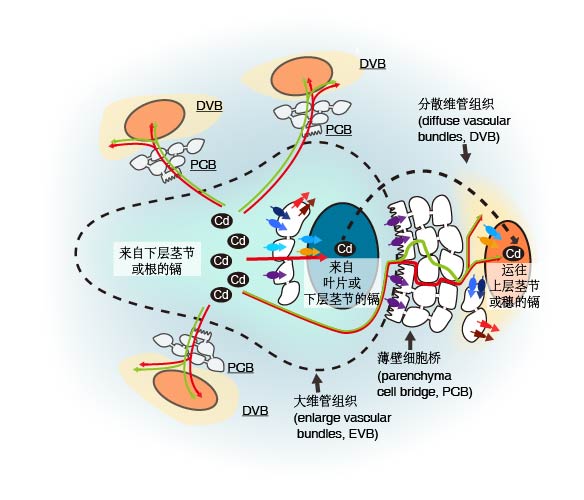
Key words: rice; cadmium; accumulation; physiological mechanisms; molecular mechanisms
Luyao Wang , Jian Chen , Shouqing Zhao , Huili Yan , Wenxiu Xu , Ruoxi Liu , Mi Ma , Yijun Yu , Zhenyan He . Research Progress of the Physiological and Molecular Mechanisms of Cadmium Accumulation in Rice[J]. Chinese Bulletin of Botany, 2022 , 57(2) : 236 -249 . DOI: 10.11983/CBB21222
| [1] | 安婷婷, 黄帝, 王浩, 张一, 陈应龙 (2021). 植物响应镉胁迫的生理生化机制研究进展. 植物学报 56, 347-362. |
| [2] | 郭韬, 余泓, 邱杰, 李家洋, 韩斌, 林鸿宣 (2019). 中国水稻遗传学研究进展与分子设计育种. 中国科学: 生命科学 49, 1185-1212. |
| [3] | 黄新元, 赵方杰 (2018). 植物防御素调控水稻镉积累的新机制. 植物学报 53, 451-455. |
| [4] | 贾沛菡 (2019). 水稻对镉的吸收受不同介质条件与生育期的影响及其与籽粒积累镉的关系. 硕士论文. 杭州: 浙江大学. pp. 1-103. |
| [5] | 李铭红, 李侠, 宋瑞生 (2008). 受污农田中农作物对重金属镉的富集特征研究. 中国生态农业学报 16, 675-679. |
| [6] | 李婷, 胡敏骏, 徐君, 蒋玉根, 闫慧莉, 虞轶俊, 何振艳 (2021). 镉低积累水稻品种选育研究进展. 中国农业科技导报 23(11), 36-46. |
| [7] | 刘婷 (2017). 镉在不同基因型水稻根系的分布及转运特征. 硕士论文. 杭州: 浙江大学. pp. 1-85. |
| [8] | 刘维涛, 周启星 (2010). 重金属污染预防品种的筛选与培育. 生态环境学报 19, 1452-1458. |
| [9] | 马卉, 焦小雨, 许学, 李娟, 倪大虎, 许蓉芳, 王钰, 汪秀峰 (2020). 水稻重金属镉代谢的生理和分子机制研究进展. 作物杂志 (1), 1-8. |
| [10] | 潘晨阳, 叶涵斐, 周维永, 王盛, 李梦佳, 路梅, 李三峰, 朱旭东, 王跃星, 饶玉春, 戴高兴 (2021). 水稻籽粒镉积累QTL定位及候选基因分析. 植物学报 56, 25-32. |
| [11] | 王宝祥, 谭明普, 刘艳, 徐大勇, 郑青松, 赵海燕, 张杰 (2020). 水稻转录因子OsNAC3在提高植物耐镉能力中的应用. 中国专利, CN111041035A. 2020-04-21. |
| [12] | 王欣梅, 肖革新, 曹贤文, 梁进军, 吴少伟 (2019). 湖南省大米中镉污染风险监测现状分析及应对策略. 环境卫生学杂志 9, 396-400, 404. |
| [13] | 肖美秀, 林文雄, 陈祥旭, 梁义元 (2006). 镉在水稻体内的分配规律与水稻镉耐性的关系. 中国农学通报 22, 379-381. |
| [14] | 徐晶晶, 吴波, 张玲妍, 郭书海, 李刚, 李凤梅 (2016). 基于贝叶斯方法的湖南湘潭稻米Cd超标风险评估. 应用生态学报 27, 3221-3227. |
| [15] | 严勋, 唐杰, 李冰, 王昌全, 徐强, 蔡欣, 付铄岚 (2019). 不同水稻品种对镉积累的差异及其与镉亚细胞分布的关系. 生态毒理学报 14(5), 244-256. |
| [16] | 杨居荣, 贺建群, 黄翌, 蒋婉茹 (1994). 农作物Cd耐性的种内和种间差异I. 种间差. 应用生态学报 5, 192-196. |
| [17] | 张蕾, 吴隆坤, 李博骞, 吴思, 王健欣 (2017). 农作物镉积累的品种差异及其机理研究进展. 北方园艺 (2), 184-190. |
| [18] | 周静, 杨洋, 孟桂元, 马国辉, 陈艳艳 (2018). 不同镉污染土壤下水稻镉富集与转运效率. 生态学杂志 37, 89-94. |
| [19] | Ansarypour Z, Shahpiri A (2017). Heterologous expression of a rice metallothionein isoform (OsMTI-1b) in Saccharomyces cerevisiae enhances cadmium, hydrogen peroxide and ethanol tolerance. Braz J Microbiol 48, 537-543. |
| [20] | Arthur EE, Crews EH, Morgan EC (2000). Optimizing plant genetic strategies for minimizing environmental contamination in the food chain. Int J Phytoremediat 2, 1-21. |
| [21] | Bari MA, El-Shehawi AM, Elseehy MM, Naheen NN, Rahman MM, Kabir AH (2021). Molecular characterization and bioinformatics analysis of transporter genes associated with Cd-induced phytotoxicity in rice (Oryza sativa L.). Plant Physiol Biochem 167, 438-448. |
| [22] | Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S (2002). Cloning an iron-regulated metal transporter from rice. J Exp Bot 53, 1677-1682. |
| [23] | Chmielowska-Bak J, Gzyl J, Rucinska-Sobkowiak R, Arasimowicz-Jelonek M, Deckert J (2014). The new insights into cadmium sensing. Front Plant Sci 5, 245. |
| [24] | Clemens S (2019). Safer food through plant science: reducing toxic element accumulation in crops. J Exp Bot 70, 5537-5557. |
| [25] | Conn S, Gilliham M (2010). Comparative physiology of elemental distributions in plants. Ann Bot 105, 1081-1102. |
| [26] | Das N, Bhattacharya S, Bhattacharyya S, Maiti MK (2017). Identification of alternatively spliced transcripts of rice phytochelatin synthase 2 gene OsPCS2 involved in mitigation of cadmium and arsenic stresses. Plant Mol Biol 94, 167-183. |
| [27] | Fu S, Lu YS, Zhang X, Yang GZ, Chao D, Wang ZG, Shi MX, Chen JG, Chao DY, Li RB, Ma JF, Xia JX (2019). The ABC transporter ABCG36 is required for cadmium tolerance in rice. J Exp Bot 70, 5909-5918. |
| [28] | Fujimaki S, Suzui N, Ishioka NS, Kawachi N, Ito S, Chino M, Nakamura SI (2010). Tracing cadmium from culture to spikelet: noninvasive imaging and quantitative characterization of absorption, transport, and accumulation of cadmium in an intact rice plant. Plant Physiol 152, 1796- 1806. |
| [29] | Gu Y, Wang P, Zhang S, Dai J, Chen HP, Lombi E, Howard DL, Van Der Ent A, Zhao FJ, Kopittke PM (2020). Chemical speciation and distribution of cadmium in rice grain and implications for bioavailability to humans. Environ Sci Technol 54, 12072-12080. |
| [30] | Hamid Y, Tang L, Yaseen M, Hussain B, Zehra A, Aziz MZ, He ZL, Yang XE (2019). Comparative efficacy of organic and inorganic amendments for cadmium and lead immobilization in contaminated soil under rice-wheat cropping system. Chemosphere 214, 259-268. |
| [31] | Hao XH, Zeng M, Wang J, Zeng ZW, Dai JL, Xie ZJ, Yang YZ, Tian LF, Chen LB, Li DP (2018). A node-expressed transporter OsCCX2 is involved in grain cadmium accumulation of rice. Front Plant Sci 9, 476. |
| [32] | Hayashi S, Tanikawa H, Kuramata M, Abe T, Ishikawa S (2020). Domain exchange between Oryza sativa phytochelatin synthases reveals a region that determines responsiveness to arsenic and heavy metals. Biochem Biophys Res Commun 523, 548-553. |
| [33] | Hu SB, Shinwari KI, Song YXR, Xia JX, Xu H, Du BB, Luo L, Zheng LQ (2021). OsNAC300 positively regulates cadmium stress responses and tolerance in rice roots. Agronomy 11, 95. |
| [34] | Hu SB, Yu Y, Chen QH, Mu GM, Shen ZG, Zheng LQ (2017). OsMYB45 plays an important role in rice resistance to cadmium stress. Plant Sci 264, 1-8. |
| [35] | Ishikawa S, Ishimaru Y, Igura M, Kuramata M, Abe T, Senoura T, Hase Y, Arao T, Nishizawa NK, Nakanishi H (2012). Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low- cadmium rice. Proc Natl Acad Sci USA 109, 19166-19171. |
| [36] | Ishikawa S, Suzui N, Ito-Tanabata S, Ishii S, Igura M, Abe T, Kuramata M, Kawachi N, Fujimaki S (2011). Real- time imaging and analysis of differences in cadmium dynamics in rice cultivars (Oryza sativa) using positron- emitting 107Cd tracer. BMC Plant Biol 11, 172. |
| [37] | Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J 45, 335-346. |
| [38] | Ishimaru Y, Takahashi R, Bashir K, Shimo H, Senoura T, Sugimoto K, Ono K, Yano M, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK (2012). Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Sci Rep 2, 286. |
| [39] | Kashiwagi T, Shindoh K, Hirotsu N, Ishimaru K (2009). Evidence for separate translocation pathways in determining cadmium accumulation in grain and aerial plant parts in rice. BMC Plant Biol 9, 8. |
| [40] | Kavitha PG, Kuruvilla S, Mathew MK (2015). Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.). Plant Physiol Biochem 97, 165-174. |
| [41] | Kuramata M, Masuya S, Takahashi Y, Kitagawa E, Inoue C, Ishikawa S, Youssefian S, Kusano T (2009). Novel cysteine-rich peptides from Digitaria ciliaris and Oryza sativa enhance tolerance to cadmium by limiting its cellular accumulation. Plant Cell Physiol 50, 106-117. |
| [42] | Li F, Wang JH, Xu L, Wang SX, Zhou MH, Yin JW, Lu AX (2018). Rapid screening of cadmium in rice and identification of geographical origins by spectral method. Int J Environ Res Public Health 15, 312. |
| [43] | Li H, Luo N, Li YW, Cai QY, Li HY, Mo CH, Wong MH (2017). Cadmium in rice: transport mechanisms, influencing factors, and minimizing measures. Environ Pollut 224, 622-630. |
| [44] | Liu CL, Gao ZY, Shang LG, Yang CH, Ruan BP, Zeng DL, Guo LB, Zhao FJ, Huang CF, Qian Q (2020). Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between Indica and Japonica rice. J Integr Plant Biol 62, 314-329. |
| [45] | Liu XS, Feng SJ, Zhang BQ, Wang MQ, Cao HW, Rono JK, Chen X, Yang ZM (2019). OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol 19, 283. |
| [46] | Luo JS, Huang J, Zeng DL, Peng JS, Zhang GB, Ma HL, Guan Y, Yi HY, Fu YL, Han B, Lin HX, Qian Q, Gong JM (2018). A defensin-like protein drives cadmium efflux and allocation in rice. Nat Commun 9, 645. |
| [47] | Lux A, Martinka M, Vaculik M, White PJ (2011). Root responses to cadmium in the rhizosphere: a review. J Exp Bot 62, 21-37. |
| [48] | Malekzadeh R, Shahpiri A (2017). Independent metal- thiolate cluster formation in C-terminal Cys-rich region of a rice type 1 metallothionein isoform. Int J Biol Macromol 96, 436-441. |
| [49] | Mao P, Zhuang P, Li F, McBride MB, Ren WD, Li YX, Li YW, Mo H, Fu HY, Li ZA (2019). Phosphate addition diminishes the efficacy of wollastonite in decreasing Cd uptake by rice (Oryza sativa L.) in paddy soil. Sci Total Environ 687, 441-450. |
| [50] | Miyadate H, Adachi S, Hiraizumi A, Tezuka K, Nakazawa N, Kawamoto T, Katou K, Kodama I, Sakurai K, Takahashi H, Satoh-Nagasawa N, Watanabe A, Fujimura T, Akagi H (2011). OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol 189, 190-199. |
| [51] | Nakanishi H, Ogawa I, Ishimaru Y, Mori S, Nishizawa NK (2006). Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci Plant Nutr 52, 464-469. |
| [52] | Nezhad RM, Shahpiri A, Mirlohi A (2013). Heterologous expression and metal-binding characterization of a type 1 metallothionein isoform (OsMTI-1b) from rice (Oryza sativa). Protein J 32, 131-137. |
| [53] | Nocito FF, Lancilli C, Dendena B, Lucchini G, Sacchi GA (2011). Cadmium retention in rice roots is influenced by cadmium availability, chelation and translocation. Plant Cell Environ 34, 994-1008. |
| [54] | Oda K, Otani M, Uraguchi S, Akihiro T, Fujiwara T (2011). Rice ABCG43 is Cd inducible and confers Cd tolerance on yeast. Biosci Biotechnol Biochem 75, 1211-1213. |
| [55] | Qi XL, Tam NFY, Li WC, Ye ZH (2020). The role of root apoplastic barriers in cadmium translocation and accumulation in cultivars of rice (Oryza sativa L.) with different Cd-accumulating characteristics. Environ Pollut 264, 114736. |
| [56] | Rodda MS, Li G, Reid RJ (2011). The timing of grain Cd accumulation in rice plants: the relative importance of remobilisation within the plant and root Cd uptake post- flowering. Plant Soil 347, 105-114. |
| [57] | Sasaki A, Yamaji N, Mitani-Ueno N, Kashino M, Ma JF (2015). A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J 84, 374-384. |
| [58] | Sebastian A, Prasad MNV (2015). Operative photo assimilation associated proteome modulations are critical for iron-dependent cadmium tolerance in Oryza sativa L. Pro- toplasma 252, 1375-1386. |
| [59] | Sebastian A, Prasad MNV (2016). Iron plaque decreases cadmium accumulation in Oryza sativa L. and serves as a source of iron. Plant Biol 18, 1008-1015. |
| [60] | Shim D, Hwang JU, Lee J, Lee S, Choi Y, An G, Martinoia E, Lee Y (2009). Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 21, 4031-4043. |
| [61] | Shimo H, Ishimaru Y, An G, Yamakawa T, Nakanishi H, Nishizawa NK (2011). Low cadmium (LCD), a novel gene related to cadmium tolerance and accumulation in rice. J Exp Bot 62, 5727-5734. |
| [62] | Song WY, Lee HS, Jin SR, Ko D, Martinoia E, Lee Y, An G, Ahn SN (2015). Rice PCR1 influences grain weight and Zn accumulation in grains. Plant Cell Environ 38, 2327-2339. |
| [63] | Song Y, Wang Y, Mao WF, Sui HX, Yong L, Yang DJ, Jiang DG, Zhang L, Gong YY (2017). Dietary cadmium exposure assessment among the Chinese population. PLoS One 12, e0177978. |
| [64] | Takahashi R, Ishimaru Y, Nakanishi H, Nishizawa NK (2011a). Role of the iron transporter OsNRAMP1 in cadmium uptake and accumulation in rice. Plant Signal Behav 6, 1813-1816. |
| [65] | Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK (2011b). The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J Exp Bot 62, 4843-4850. |
| [66] | Takahashi R, Ishimaru Y, Shimo H, Ogo Y, Senoura T, Nishizawa NK, Nakanishi H (2012). The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ 35, 1948-1957. |
| [67] | Tan LT, Qu MM, Zhu YX, Peng C, Wang JR, Gao DY, Chen CY (2020). ZINC TRANSPORTER5 and ZINC TRANSPORTER9 function synergistically in Zinc/Cadmium uptake. Plant Physiol 183, 1235-1249. |
| [68] | Tan LT, Zhu YX, Fan T, Peng C, Wang JR, Sun L, Chen CY (2019). OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem Biophys Res Commun 512, 112-118. |
| [69] | Tanaka K, Fujimaki S, Fujiwara T, Yoneyama T, Hayashi H (2003). Cadmium concentrations in the phloem sap of rice plants (Oryza sativa L.) treated with a nutrient solution containing cadmium. Soil Sci Plant Nutr 49, 311-313. |
| [70] | Tanaka K, Fujimaki S, Fujiwara T, Yoneyama T, Hayashi H (2007). Quantitative estimation of the contribution of the phloem in cadmium transport to grains in rice plants (Oryza sativa L.). Soil Sci Plant Nutr 53, 72-77. |
| [71] | Tanaka N, Uraguchi S, Kajikawa M, Saito A, Ohmori Y, Fujiwara T (2018). A rice PHD-finger protein OsTITANIA, is a growth regulator that functions through elevating expression of transporter genes for multiple metals. Plant J 96, 997-1006. |
| [72] | Tang L, Mao BG, Li YK, Lv QM, Zhang LP, Chen CY, He HJ, Wang WP, Zeng XF, Shao Y, Pan YL, Hu YY, Peng Y, Fu XQ, Li HQ, Xia ST, Zhao BR (2017). Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci Rep 7, 14438. |
| [73] | Tefera W, Liu T, Lu LL, Ge J, Webb SM, Seifu W, Tian SK (2020). Micro-XRF mapping and quantitative assessment of Cd in rice (Oryza sativa L.) roots. Ecotoxicol Environ Saf 193, 110245. |
| [74] | Ueno D, Yamaji N, Kono I, Huang CF, Ando T, Yano M, Ma JF (2010). Gene limiting cadmium accumulation in rice. Proc Natl Acad Sci USA 107, 16500-16505. |
| [75] | Uraguchi S, Fujiwara T (2013). Rice breaks ground for cadmium-free cereals. Curr Opin Plant Biol 16, 328-334. |
| [76] | Uraguchi S, Kamiya T, Sakamoto T, Kasai K, Sato Y, Nagamura Y, Yoshida A, Kyozuka J, Ishikawa S, Fujiwara T (2011). Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc Natl Acad Sci USA 108, 20959-20964. |
| [77] | Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S (2009). Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J Exp Bot 60, 2677-2688. |
| [78] | Uraguchi S, Tanaka N, Hofmann C, Abiko K, Ohkama- Ohtsu N, Weber M, Kamiya T, Sone Y, Nakamura R, Takanezawa Y, Kiyono M, Fujiwara T, Clemens S (2017). Phytochelatin synthase has contrasting effects on cadmium and arsenic accumulation in rice grains. Plant Cell Physiol 58, 1730-1742. |
| [79] | Wang FJ, Tan HF, Han JH, Zhang YT, He X, Ding YF, Chen ZX, Zhu C (2019). A novel family of PLAC8 motif-containing/PCR genes mediates Cd tolerance and Cd accumulation in rice. Environ Sci Eur 31, 82. |
| [80] | Wang ME, Chen WP, Peng C (2016). Risk assessment of Cd polluted paddy soils in the industrial and township areas in Hunan, Southern China. Chemosphere 144, 346- 351. |
| [81] | Xiong WT, Wang P, Yan TZ, Cao BB, Xu J, Liu DF, Luo MZ (2018). The rice "fruit-weight 2.2-like" gene family member OsFWL4 is involved in the translocation of cadmium from roots to shoots. Planta 247, 1247-1260. |
| [82] | Yamaguchi N, Ishikawa S, Abe T, Baba K, Arao T, Terada Y (2012). Role of the node in controlling traffic of cadmium, zinc, and manganese in rice. J Exp Bot 63, 2729- 2737. |
| [83] | Yamaji N, Ma JF (2014). The node, a hub for mineral nutrient distribution in graminaceous plants. Trends Plant Sci 19, 556-563. |
| [84] | Yamaji N, Ma JF (2017). Node-controlled allocation of mine- ral elements in Poaceae. Curr Opin Plant Biol 39, 18-24. |
| [85] | Yamaji N, Xia JX, Mitani-Ueno N, Yokosho K, Ma JF (2013). Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2. Plant Physiol 162, 927-939. |
| [86] | Yan HL, Xu WX, Xie JY, Gao YW, Wu LL, Sun L, Feng L, Chen X, Zhang T, Dai CH, Li T, Lin XN, Zhang ZY, Wang XQ, Li FM, Zhu XY, Li JJ, Li ZC, Chen CY, Ma M, Zhang HL, He ZY (2019). Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies. Nat Commun 10, 2562. |
| [87] | Yuan LY, Yang SG, Liu BX, Zhang M, Wu KQ (2012). Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep 31, 67-79. |
| [88] | Zhao FJ, Huang XY (2018). Cadmium phytoremediation: Call Rice CAL1. Mol Plant 11, 640-642. |
| [89] | Zhao FJ, Wang P (2020). Arsenic and cadmium accumulation in rice and mitigation strategies. Plant Soil 446, 1-21. |
| [90] | Zhao JL, Yang W, Zhang SH, Yang TF, Liu Q, Dong JF, Fu H, Mao XX, Liu B (2018). Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection. Rice 11, 61. |
| [91] | Zheng X, Chen L, Li XF (2018). Arabidopsis and rice showed a distinct pattern in ZIPs genes expression profile in response to Cd stress. Bot Stud 59, 22. |
/
| 〈 |
|
〉 |