

QTL Mapping and Candidate Gene Analysis of Vitamin E in Rice Grain
Received date: 2021-12-12
Accepted date: 2022-01-20
Online published: 2022-01-20
Vitamin E (VE) is an important index of rice nutritional quality. Rice (Oryza sativa) is the most widely planted crop in China. To increase rice VE content is a convenient and effective way to improve its nutrient composition. In this study, 120 recombinant inbred line (RIL) populations were constructed with indica rice HZ as male parent and japonica rice Nekken2 as female parent. The content of vitamin E isomer in the population was determined by high performance liquid chromatography (HPLC). Quantitative trait loci (QTL) mapping was carried out based on the constructed high-density molecular genetic map. After pedigree analysis, 122 VE related QTLs were identified, which were distributed on 12 chromosomes. Some loci have high LOD scores, such as qT3α/to2-1 with 10.32 LOD score, qT3α2-1 of 9.91 LOD score. There are multiple major QTLs associated with the content of different conformations, and some VE biosynthesis genes are within the intervals, e.g., OsGGR1, OsGGR2, OsTC and OsγTMT. The expression of these genes were analyzed by qRT-PCR. The results showed that their expression in HZ was significantly higher than that in Nekken2. It is speculated that the high expression of these genes is the reason of why tocopherol and tocotrienol in HZ are higher than in Nekken2. The number of QTLs and the scores of LOD are large, which provides a molecular basis for further screening and cultivating rice varieties with high VE content also conductive to clarify the molecular regulation mechanism of rice VE biosynthesis through QTLs.
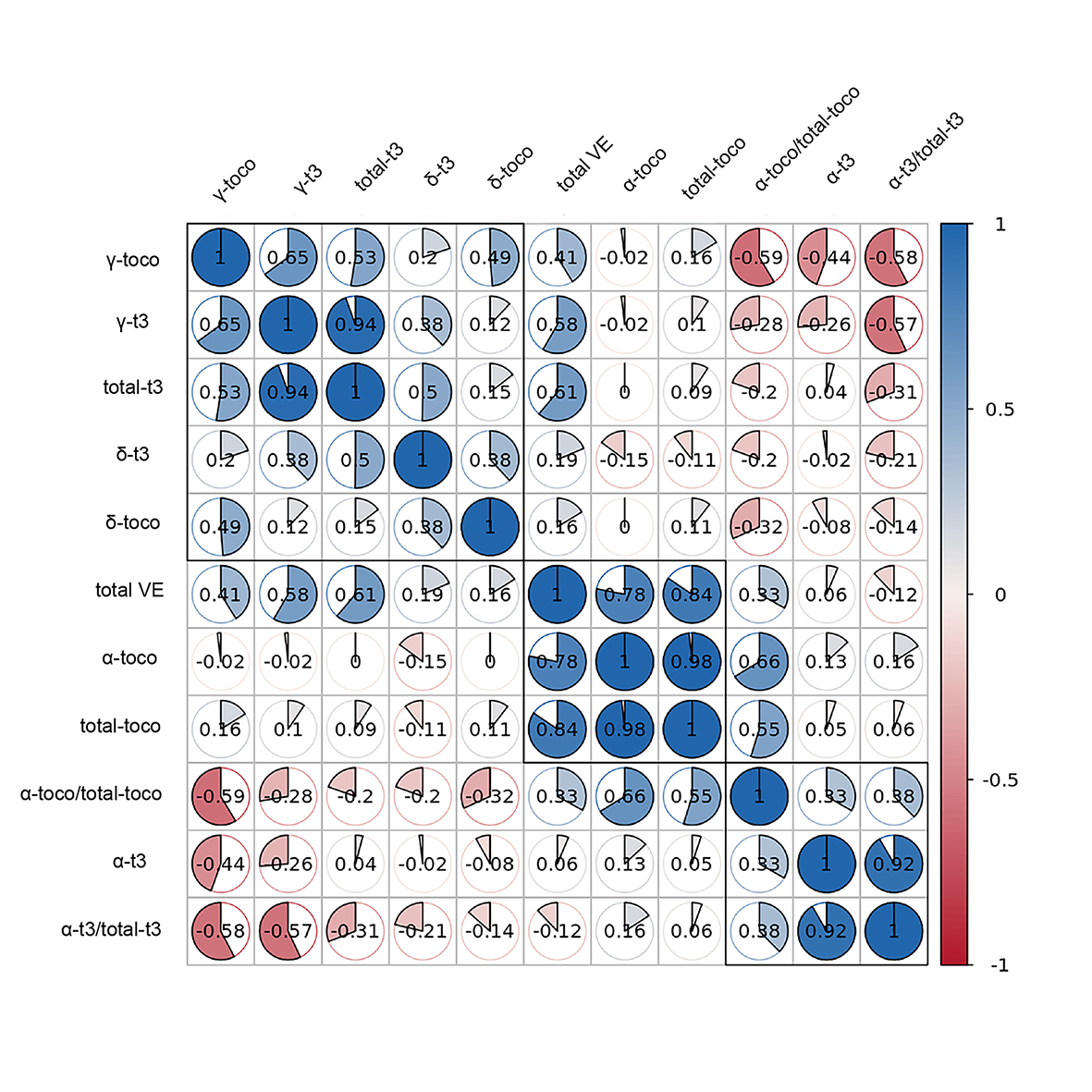
Key words: rice; vitamin E; QTL mapping; candidate genes; rice quality
Hanfei Ye , Wenjing Yin , Yian Guan , Kairu Yang , Qianyu Chen , Shuying Yu , Xudong Zhu , Dedong Xin , Wei Zhang , Yuexing Wang , Yuchun Rao . QTL Mapping and Candidate Gene Analysis of Vitamin E in Rice Grain[J]. Chinese Bulletin of Botany, 2022 , 57(2) : 157 -170 . DOI: 10.11983/CBB21216
| [1] | 江良荣, 李义珍, 王侯聪, 黄育民 (2004). 稻米营养品质的研究现状及分子改良途径. 分子植物育种 1, 112-120. |
| [2] | 李春梅 (2019). 两个水稻黄绿叶突变基因的图位克隆及功能分析. 博士论文. 成都: 四川农业大学. pp. 24-84. |
| [3] | 潘晨阳, 叶涵斐, 周维永, 王盛, 李梦佳, 路梅, 李三峰, 朱旭东, 王跃星, 饶玉春, 戴高兴 (2021a). 水稻籽粒镉积累QTL定位及候选基因分析. 植物学报 56, 25-32. |
| [4] | 潘晨阳, 张月, 林晗, 陈芊羽, 杨凯如, 姜嘉骥, 李梦佳, 芦涛, 王珂欣, 路梅, 王盛, 叶涵斐, 饶玉春, 胡海涛 (2021b). 水稻叶片水势的QTL定位与候选基因分析. 植物学报 56, 275-283. |
| [5] | 潘卫东, 李晓峰, 陈双燕, 刘公社 (2006). 植物维生素E合成相关酶基因的克隆及其在体内功能研究进展. 植物学通报 23, 68-77. |
| [6] | 饶玉春, 戴志俊, 朱怡彤, 姜嘉骥, 马若盈, 王予烨, 王跃星 (2020a). 水稻抗干旱胁迫的研究进展. 浙江师范大学学报(自然科学版) 43, 417-429. |
| [7] | 饶玉春, 林晗, 肖飒清, 吴仪, 张月, 王盛, 胡佳青, 薛大伟 (2020b). 水稻耐淹性的QTL定位与候选基因分析. 浙江师范大学学报(自然科学版) 43, 312-319. |
| [8] | 王迪 (2017). 水稻生育酚合成基因SGD1的图位克隆与功能分析. 博士论文. 南京: 南京农业大学. pp. 35-58. |
| [9] | 尹雪斌, 赵其国, 印遇龙, 陈清清, 王张民, 宋佳平, 王晓丽, 林锦钰, 王莉莉, 刘晓航, 张泽洲, 张宁 (2022). 功能农业关键科学问题研究进展与“十四五”发展建议. 科学通报 67, 497-510. |
| [10] | 章薇 (2017). 拟南芥叶绿素代谢与维生素E合成代谢的关系研究. 博士论文. 武汉: 华中农业大学. pp. 1-23. |
| [11] | 章秀福, 王丹英, 方福平, 曾衍坤, 廖西元 (2005). 中国粮食安全和水稻生产. 农业现代化研究 26, 85-88. |
| [12] | 周纯, 焦然, 胡萍, 林晗, 胡娟, 徐娜, 吴先美, 饶玉春, 王跃星 (2019). 水稻早衰突变体LS-es1的基因定位及候选基因分析. 植物学报 54, 606-619. |
| [13] | 周浩, 夏朵, 赵达, 李艳华, 李平波, 吴边, 高冠军, 张庆路, 何予卿 (2020). 稻米食味品质的遗传研究进展. 见:第十九届中国作物学会学术年会论文摘要集. 武汉: 中国作物学会. pp. 56. |
| [14] | Chaudhary N, Khurana P (2009). Vitamin E biosynthesis genes in rice: molecular characterization, expression profiling and comparative phylogenetic analysis. Plant Sci 177, 479-491. |
| [15] | Hosomi A, Arita M, Sato Y, Kiyose C, Ueda T, Igarashi O, Arai H, Inoue K (1997). Affinity for α-tocopherol transfer protein as a determinant of the biological activities of vitamin E analogs. FEBS Lett 409, 105-108. |
| [16] | Hwang JE, Ahn JW, Kwon SJ, Kim JB, Kim SH, Kang SY, Kim DS (2014). Selection and molecular characterization of a high tocopherol accumulation rice mutant line induced by gamma irradiation. Mol Biol Rep 41, 7671-7681. |
| [17] | Kimura E, Abe T, Murata K, Kimura T, Otoki Y, Yoshida T, Miyazawa T, Nakagawa K (2018). Identification of OsGGR2, a second geranylgeranyl reductase involved in α-tocopherol synthesis in rice. Sci Rep 8, 1870. |
| [18] | Kruk J, Strzałka K (1995). Occurrence and function of α- tocopherol quinone in plants. J Plant Physiol 145, 405-409. |
| [19] | Li Y, Wang ZN, Sun XF, Tang KX (2008). Current opinions on the functions of tocopherol based on the genetic manipulation of tocopherol biosynthesis in plants. J Integr Plant Biol 50, 1057-1069. |
| [20] | Luo W, Huan Q, Xu YY, Qian WF, Chong K, Zhang JY (2021). Integrated global analysis reveals a vitamin E- vitamin K1 sub-network, downstream of COLD1, underlying rice chilling tolerance divergence. Cell Rep 36(3), 58- 76. |
| [21] | McCouch SR, Cho YC, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997). Report on QTL nomenclature. Rice Genet Newsl 14, 11-13. |
| [22] | Sekine D, Murata K, Kimura T, Nakagawa K, Miyazawa T (2016). Identification of a genetic factor required for high γ-isoform concentration in rice vitamin E. J Agric Food Chem 64, 9368-9373. |
| [23] | Sookwong P, Murata K, Nakagawa K, Shibata A, Kimura T, Yamaguchi M, Kojima Y, Miyazawa T (2009). Cross- fertilization for enhancing tocotrienol biosynthesis in rice plants and QTL analysis of their F2 progenies. J Agric Food Chem 57, 4620-4625. |
| [24] | Tucker JM, Townsend DM (2005). Alpha-tocopherol: roles in prevention and therapy of human disease. Biomed Pharmacother 59, 380-387. |
| [25] | Wang X, Song YE, Li JY (2013). High expression of tocochromanol biosynthesis genes increases the vitamin E level in a new line of giant embryo rice. J Agric Food Chem 61, 5860-5869. |
| [26] | Wang XQ, Yoon MY, He Q, Kim TS, Tong W, Choi BW, Lee YS, Park YJ (2015). Natural variations in OsγTMT contribute to diversity of the α-tocopherol content in rice. Mol Genet Genomics 290, 2121-2135. |
| [27] | Wang YX, Shang LG, Yu H, Zeng LJ, Hu J, Ni S, Rao YC, Li SF, Chu JF, Meng XB, Wang L, Hu P, Yan JJ, Kang SJ, Qu MH, Lin H, Wang T, Wang Q, Hu XM, Chen HQ, Wang B, Gao ZY, Guo LB, Zeng DL, Zhu XD, Xiong GS, Li JY, Qian Q (2020). A strigolactone biosynthesis gene contributed to the green revolution in rice. Mol Plant 13, 923-932. |
| [28] | Yang WY, Cahoon RE, Hunter SC, Zhang CY, Han JX, Borgschulte T, Cahoon EB (2011). Vitamin E biosynthesis: functional characterization of the monocot homogenti- sate geranylgeranyl transferase. Plant J 65, 206-217. |
| [29] | Zhang CY, Cahoon RE, Hunter SC, Chen M, Han JX, Cahoon EB (2013). Genetic and biochemical basis for alternative routes of tocotrienol biosynthesis for enhanced vitamin E antioxidant production. Plant J 73, 628-639. |
| [30] | Zhang XN, Li PB, Yun P, Wang LQ, Gao GJ, Zhang QL, Luo LJ, Zhang CY, He YQ (2019). Genetic dissection of vitamin E content in rice (Oryza sativa) grains using recombinant inbred lines derived from a cross between ‘Zhenshan97B’ and ‘Nanyangzhan’. Plant Breed 138, 820- 829. |
/
| 〈 |
|
〉 |