

Regulatory Mechanisms of the Plant CBL-CIPK Signaling System in Response to Abiotic Stress
Received date: 2021-01-29
Accepted date: 2021-04-19
Online published: 2021-04-21
Calcineurin B-like proteins (CBLs) and their CBL-interacting protein kinases (CIPKs) are important regulatory network in response to abiotic stresses. The CBL-CIPK system senses and decodes Ca2+-signals through phosphorylation to regulate plant response to abiotic stresses. In this review, the basic structures of CBLs and CIPKs, and their phosphorylation on different substrates, as well as regulatory mechanisms of plants in response to abiotic stresses were summarized. We also put forward a perspective on the future research directions of CBLs and CIPKs, as well as their potential applications in genetic improvement of crops for stress tolerance.
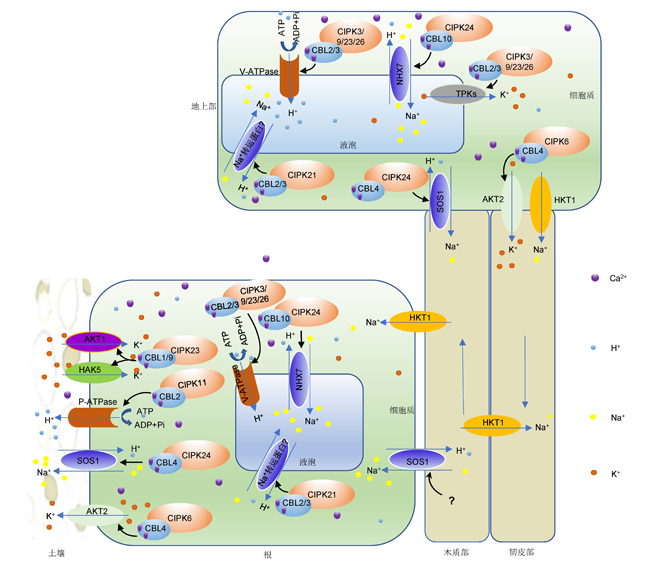
Key words: abiotic stresses; calcium signaling; CBL-CIPK; ion channels; phosphorylation
Lingling Xie , Jinlong Wang , Guoqiang Wu . Regulatory Mechanisms of the Plant CBL-CIPK Signaling System in Response to Abiotic Stress[J]. Chinese Bulletin of Botany, 2021 , 56(5) : 614 -626 . DOI: 10.11983/CBB21024
| [1] | 沈金秋, 郑仲仲, 潘伟槐, 潘建伟 (2014). 植物CBL-CIPK信号系统的功能及其作用机理. 植物生理学报 50, 641-650. |
| [2] | 伍国强, 水清照, 冯瑞军 (2017). 植物K+通道AKT1的研究进展. 植物学报 52, 225-234. |
| [3] | 张和臣, 尹伟伦, 夏新莉 (2007). 非生物逆境胁迫下植物钙信号转导的分子机制. 植物学通报 24, 114-122. |
| [4] | Alemán F, Nieves-Cordones M, Martínez V, Rubio F (2011). Root K+ acquisition in plants: the Arabidopsis thaliana model. Plant Cell Physiol 52, 1603-1612. |
| [5] | Almeida DM, Oliveira MM, Saibo NJM (2017). Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet Mol Biol 40, 326-345. |
| [6] | Aslam M, Fakher B, Jakada BH, Zhao LH, Cao SJ, Cheng Y, Qin Y (2019). Genome-wide identification and expression profiling of CBL-CIPK gene family in pineapple ( Ananas comosus) and the role of AcCBL1 in abiotic and biotic stress response. Biomolecules 9, 293. |
| [7] | Batistič O, Waadt R, Steinhorst L, Held K, Kudla J (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61, 211-222. |
| [8] | Beckmann L, Edel KH, Batistič O, Kudla J (2016). A calcium sensor-protein kinase signaling module diversified in plants and is retained in all lineages of Bikonta species. Sci Rep 6, 31645. |
| [9] | Behera S, Long Y, Schmitz-Thom I, Wang XP, Zhang CX, Li H, Steinhorst L, Manishankar P, Ren XL, Offenborn JN, Wu WH, Kudla J, Wang Y (2017). Two spatially and temporally distinct Ca2+ signals convey Arabidopsis thaliana responses to K+ deficiency. New Phytol 213, 739-750. |
| [10] | Bender KW, Zielinski RE, Huber SC (2018). Revisiting paradigms of Ca2+ signaling protein kinase regulation in plants. Biochem J 475, 207-223. |
| [11] | Campbell MT, Bandillo N, Al Shiblawi FRA, Sharma S, Liu K, Du Q, Schmitz AJ, Zhang C, Véry A, Lorenz AJ, Walia H (2017). Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PLoS Genet 13, e1006823. |
| [12] | Chaves-Sanjuan A, Sanchez-Barrena MJ, Gonzalez-Rubio JM, Moreno M, Ragel P, Jimenez M, Pardob JM, Martinez-Ripoll M, Quintero FJ, Albert A (2014). Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress. Proc Natl Acad Sci USA 111, E4532-E4541. |
| [13] | Chen XF, Gu ZM, Xin DD, Hao L, Liu CJ, Huang J, Ma BJ, Zhang HS (2011). Identification and characterization of putative CIPK genes in maize. J Genet Genomics 38, 77-87. |
| [14] | Chu LC, Offenborn JN, Steinhorst L, Wu XN, Xi L, Li Z, Jacquot A, Lejay L, Kudla J, Schulze WX (2021). Plasma membrane calcineurin B-like calcium-ion sensor proteins function in regulating primary root growth and nitrate uptake by affecting global phosphorylation patterns and microdomain protein distribution. New Phytol 229, 2223-2237. |
| [15] | Cui XY, Du YT, Fu JD, Yu TF, Wang TC, Chen M, Chen J, Ma YZ, Xu ZS (2018). Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses. BMC Plant Biol 18, 93-106. |
| [16] | Dong QY, Bai BW, Almutairi BO, Kudla J (2021). Emerging roles of the CBL-CIPK calcium signaling network as key regulatory hub in plant nutrition. J Plant Physiol 257, 153335. |
| [17] | Guo Y, Halfter U, Ishitani M, Zhu JK (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13, 1383-1400. |
| [18] | Guo Y, Xiong LM, Song CP, Gong DM, Halfter U, Zhu JK (2002). A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3, 233-244. |
| [19] | Han JP, Köster P, Drerup MM, Scholz M, Li SZ, Edel KH, Hashimoto K, Kuchitsu K, Hippler M, Kudla J (2019). Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca2+ binding. New Phytol 221, 1935-1949. |
| [20] | Hashimoto K, Eckert C, Anschütz U, Scholz M, Held K, Waadt R, Reyer A, Hippler M, Becker D, Kudla J (2012). Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J Biol Chem 287, 7956-7968. |
| [21] | Hepler PK (2005). Calcium: a central regulator of plant growth and development. Plant Cell 17, 2142-2155. |
| [22] | Ho CH, Lin SH, Hu HC, Tsay YF (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184-1194. |
| [23] | Hu HC, Wang YY, Tsay YF (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J 57, 264-278. |
| [24] | Hu W, Xia ZQ, Yan Y, Ding ZH, Tie WW, Wang LZ, Zou ML, Wei YX, Lu C, Hou XW, Wang XQ, Peng M (2015). Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Front Plant Sci 6, 914. |
| [25] | Hwang YS, Bethke PC, Cheong YH, Chang HS, Zhu T, Jones RL (2005). A gibberellin-regulated calcineurin B in rice localizes to the tonoplast and is implicated in vacuole function. Plant Physiol 138, 1347-1358. |
| [26] | Kim BG, Waadt R, Cheong YH, Pandey JK, Dominguez- Solis JR, Schültke S, Lee SC, Kudla J, Luan S (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J 52, 473-484. |
| [27] | Kimura S, Kawarazaki T, Nibori H, Michikawa M, Imai A, Kayayz H, Kuchitsu K (2013). The CBL-interacting protein kinase CIPK26 is a novel interactor of Arabidopsis NADPH oxidase AtRbohF that negatively modulates its ROS-producing activity in a heterologous expression system. J Biochem 153, 191-195. |
| [28] | Kleist TJ, Spencley AL, Luan S (2014). Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front Plant Sci 5, 187. |
| [29] | Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134, 43-58. |
| [30] | Köster P, Wallrad L, Edel KH, Faisal M, Alatar AA, Kudla J (2019). The battle of two ions: Ca2+ signaling against Na+ stress. Plant Biol 21, 39-48. |
| [31] | Kudla J, Becker D, Grill E, Hedrich R, Hippler M, Kummer U, Parniske M, Romeis T, Schumacher K (2018). Advances and current challenges in calcium signaling. New Phytol 218, 414-431. |
| [32] | Lara A, Ródenas R, Andrés Z, Martínez V, Quintero FJ, Nieves-Cordones M, Botella MA, Rubio F (2020). Arabidopsis K+ transporter HAK5-mediated high-affinity root K+ uptake is regulated by protein kinases CIPK1 and CIPK9. J Exp Bot 71, 5053-5060. |
| [33] | Léran S, Edel KH, Pervent M, Hashimoto K, Corratgé- Faillie C, Offenborn JN, Tillard P, Gojon A, Kudla J, Lacombe B (2015). Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci Signal 8, ra43. |
| [34] | Li J, Jiang MM, Li R, Yang L, Liu Y, Chen HY (2016). Identification and characterization of CBL and CIPK gene families in eggplant ( Solanum melongena L.). Mol Genet Genomics 291, 1769-1781. |
| [35] | Lin HX, Du WM, Yang YQ, Schumaker KS, Guo Y (2014). A calcium-independent activation of the Arabidopsis SOS2-like protein kinase 24 by its interacting SOS3-like calcium binding protein. Plant Physiol 164, 2197-2206. |
| [36] | Liu H, Wang YX, Li H, Teng RM, Wang Y, Zhuang J (2019). Genome-wide identification and expression analysis of calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase family genes in tea plant. DNA Cell Biol 38, 824-839. |
| [37] | Liu LT, Zheng CH, Kuang BJ, Wei LQ, Yan LF, Wang T (2016). Receptor-like kinase RUPO interacts with potassium transporters to regulate pollen tube growth and integrity in rice. PLoS Genet 12, e1006085. |
| [38] | Liu P, Duan YH, Liu C, Xue QH, Guo J, Qi T, Kang ZS, Guo J (2018). Corrigendum to: the calcium sensor TaCBL4 and its interacting protein TaCIPK5 are required for wheat resistance to stripe rust fungus. J Exp Bot 69, 5309. |
| [39] | López-Arredondo DL, Leyva-González MA, Alatorre- Cobos F, Herrera-Estrella L (2013). Biotechnology of nutrient uptake and assimilation in plants. Int J Dev Biol 57, 595-610. |
| [40] | López-Arredondo DL, Leyva-González MA, González- Morales SI, Lopez-Bucio J, Herrera-Estrella L (2014). Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol 65, 95-123. |
| [41] | Lu TT, Zhang GF, Sun LR, Wang J, Hao FS (2017). Genome-wide identification of CBL family and expression analysis of CBLs in response to potassium deficiency in cotton. Peer J 5, e3653. |
| [42] | Lyzenga WJ, Sullivan V, Liu HX, Stone SL (2017). The kinase activity of calcineurin B-like interacting protein kinase 26 (CIPK26) influences its own stability and that of the ABA-regulated ubiquitin ligase, keep on going (KEG). Front Plant Sci 8, 502. |
| [43] | Ma Q, Tang RJ, Zheng XJ, Wang SM, Luan S (2015). The calcium sensor CBL7 modulates plant responses to low nitrate in Arabidopsis. Biochem Biophys Res Commun 468, 59-65. |
| [44] | Ma X, Gai WX, Qiao YM, Ali M, Wei AM, Luo DX, Li QH, Gong ZH (2019). Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genomics 20, 775. |
| [45] | Ma X, Li QH, Yu YN, Qiao YM, Haq S, Gong ZH (2020). The CBL-CIPK pathway in plant response to stress signals. Int J Mol Sci 21, 5668. |
| [46] | Ma YC, Cheng QK, Cheng ZM, Li H, Chang YH, Lin J (2017). Identification of important physiological traits and moderators that are associated with improved salt tolerance in CBL and CIPK overexpressors through a meta- analysis. Front Plant Sci 8, 856. |
| [47] | Mahajan S, Sopory SK, Tuteja N (2006). Cloning and characterization of CBL-CIPK signaling components from a legume ( Pisum sativum). FEBS J 273, 907-925. |
| [48] | Maierhofer T, Diekmann M, Offenborn JN, Lind C, Bauer H, Hashimoto K, Al-Rasheid KAS, Luan S, Kudla J, Geiger D, Hedrich R (2014a). Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci Signal 7, ra86. |
| [49] | Maierhofer T, Lind C, Hüttl S, Scherzer S, Papenfuβ M, Simon J, Al-Rasheid KAS, Ache P, Rennenberg H, Hedrich R, Müller TD, Geiger D (2014b). A single-pore residue renders the Arabidopsis root anion channel SLAH2 highly nitrate selective. Plant Cell 26, 2554-2567. |
| [50] | Manik SMN, Shi SJ, Mao JJ, Dong LH, Su YL, Wang Q, Liu HB (2015). The calcium sensor CBL-CIPK is involved in plant’s response to abiotic stresses. Int J Genomics 2015, 493191. |
| [51] | Manishankar P, Wang N, Köster P, Alatar AA, Kudla J (2018). Calcium signaling during salt stress and in the regulation of ion homeostasis. J Exp Bot 69, 4215-4226. |
| [52] | Mao JJ, Manik S, Shi SJ, Chao JT, Jin YR, Wang Q, Liu HB (2016). Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana. Genes 7, 62-77. |
| [53] | Mo CY, Wan SM, Xia YQ, Ren N, Zhou Y, Jiang XY (2018). Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses. Front Plant Sci 9, 269. |
| [54] | Niu LL, Dong BY, Song ZH, Meng D, Fu YJ (2018). Genome-wide identification and characterization of CIPK family and analysis responses to various stresses in apple (Malus domestica). Int J Mol Sci 19, 2131. |
| [55] | Oda Y, Kobayashi NI, Tanoi K, Ma JF, Itou Y, Katsuhara M, Itou T, Horie T (2018). T-DNA tagging-based gain-of- function of OsHKT1;4 reinforces Na exclusion from leaves and stems but triggers Na toxicity in roots of rice under salt stress. Int J Mol Sci 19, 235. |
| [56] | Pandey GK, Grant JJ, Cheong YH, Kim BG, Li LG, Luan S (2008). Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol Plant 1, 238-248. |
| [57] | Parker JL, Newstead S (2014). Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507, 68-72. |
| [58] | Peck S, Mittler R (2020). Plant signaling in biotic and abiotic stress. J Exp Bot 71, 1649-1651. |
| [59] | Piao HL, Xuan YH, Park SH, Je BI, Park SJ, Park SH, Kim CM, Huang J, Wang GK, Kim MJ, Kang SM, Lee IJ, Kwon TR, Kim YH, Yeo US, Yi G, Son D, Han CD (2010). OsCIPK31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants. Mol Cells 30, 19-27. |
| [60] | Plasencia FA, Estrada Y, Flores FB, Ortíz-Atienza A, Lozano R, Egea I (2021). The Ca2+ sensor calcineurin B-like protein 10 in plants: emerging new crucial roles for plant abiotic stress tolerance. Front Plant Sci 11, 599944. |
| [61] | Ragel P, Raddatz N, Leidi EO, Quintero FJ, Pardo JM (2019). Regulation of K+ nutrition in plants. Front Plant Sci 10, 281. |
| [62] | Saito S, Hamamoto S, Moriya K, Matsuura A, Sato Y, Muto J, Noguchi H, Yamauchi S, Tozawa Y, Ueda M, Hashimoto K, Köster P, Dong QY, Held K, Kudla J, Utsumi T, Uozumi N (2018). N-myristoylation and S-acylation are common modifications of Ca2+-regulated Arabidopsis kinases and are required for activation of the SLAC1 anion channel. New Phytol 218, 1504-1521. |
| [63] | Saito S, Uozumi N (2019). Guard cell membrane anion transport systems and their regulatory components: an elaborate mechanism controlling stress-induced stomatal closure. Plants 8, 9. |
| [64] | Saito S, Uozumi N (2020). Calcium-regulated phosphorylation systems controlling uptake and balance of plant nutrients. Front Plant Sci 11, 44. |
| [65] | Sánchez-Barrena MJ, Chaves-Sanjuan A, Raddatz N, Mendoza I, Cortés Á, Gago F, González-Rubio JM, Benavente JL, Quintero FJ, Pardo JM, Albert A (2020). Recognition and activation of the plant AKT1 potassium channel by the kinase CIPK23. Plant Physiol 182, 2143-2153. |
| [66] | Sánchez-Barrena MJ, Martínez-Ripoll M, Albert A (2013). Structural biology of a major signaling network that regulates plant abiotic stress: the CBL-CIPK mediated pathway. Int J Mol Sci 14, 5734-5749. |
| [67] | Sánchez-Barrena MJ, Martínez-Ripoll M, Zhu JK, Albert A (2005). The structure of the Arabidopsis thaliana SOS3: molecular mechanism of sensing calcium for salt stress response. J Mol Biol 345, 1253-1264. |
| [68] | Sanyal SK, Kanwar P, Yadav AK, Sharma C, Kumar A, Pandey GK (2017). Arabidopsis CBL interacting protein kinase 3 interacts with ABR1, an APETALA2 domain transcription factor, to regulate ABA responses. Plant Sci 254, 48-59. |
| [69] | Sanyal SK, Mahiwal S, Nambiar DM, Pandey GK (2020). CBL-CIPK module-mediated phosphoregulation: facts and hypothesis. Biochem J 477, 853-871. |
| [70] | Sanyal SK, Pandey A, Pandey GK (2015). The CBL-CIPK signaling module in plants: a mechanistic perspective. Physiol Plant 155, 89-108. |
| [71] | Sanyal SK, Rao S, Mishra LK, Sharma M, Pandey GK (2016). Plant stress responses mediated by CBL-CIPK phosphorylation network. Enzymes 40, 31-64. |
| [72] | Sierla M, Waszczak C, Vahisalu T, Kangasjärvi J (2016). Reactive oxygen species in the regulation of stomatal movements. Plant Physiol 171, 1569-1580. |
| [73] | Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang PC, Zhu JK (2005). Role of an Arabidopsis AP2/EREBP- type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384-2396. |
| [74] | Song SJ, Feng QN, Li CL, Li E, Liu Q, Kang H, Zhang W, Zhang Y, Li S (2018). A tonoplast-associated calcium- signaling module dampens ABA signaling during stomatal movement. Plant Physiol 177, 1666-1678. |
| [75] | Srivastava AK, Shankar A, Chandran AKN, Sharma M, Jung KH, Suprasanna P, Pandey GK (2020). Emerging concepts of potassium homeostasis in plants. J Exp Bot 71, 608-619. |
| [76] | Steinhorst L, Kudla J (2013). Calcium and reactive oxygen species rule the waves of signaling. Plant Physiol 163, 471-485. |
| [77] | Straub T, Ludewig U, Neuhäuser B (2017). The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell 29, 409-422. |
| [78] | Su WH, Ren YJ, Wang DJ, Huang L, Fu XQ, Ling H, Su YC, Huang N, Tang HC, Xu LP, Que YX (2020). New insights into the evolution and functional divergence of the CIPK gene family in Saccharum. BMC Genomics 21, 868-888. |
| [79] | Sun J, Bankston JR, Payandeh J, Hinds TR, Zagotta WN, Zheng N (2014). Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73-77. |
| [80] | Sun T, Wang Y, Wang M, Li TT, Zhou Y, Wang XT, Wei SY, He GY, Yang GX (2015). Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol 15, 269. |
| [81] | Tang RJ, Liu H, Yang Y, Yang L, Gao XS, Garcia VJ, Luan S, Zhang HX (2012). Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res 22, 1650-1665. |
| [82] | Tang RJ, Wang C, Li KL, Luan S (2020). The CBL-CIPK calcium signaling network: unified paradigm from 20 years of discoveries. Trends Plant Sci 25, 604-617. |
| [83] | Tang RJ, Zhao FG, Garcia VJ, Kleista TJ, Yang L, Zhang HX, Luan S (2015). Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc Natl Acad Sci USA 112, 3134-3139. |
| [84] | Toyota M, Spencer D, Sawai-Toyota S, Wang JQ, Zhang T, Koo AJ, Howe GA, Gilroy S (2018). Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361, 1112-1115. |
| [85] | Tripathi V, Syed N, Laxmi A, Chattopadhyay D (2009). Role of CIPK6 in root growth and auxin transport. Plant Signal Behav 4, 663-665. |
| [86] | Wang XP, Zhu BP, Jiang ZH, Wang SC (2019). Calcium-mediation of jasmonate biosynthesis and signaling in plants. Plant Sci 287, 110192. |
| [87] | Wang Y, Li TT, John SJ, Chen MJ, Chang JL, Yang GX, He GY (2018). A CBL-interacting protein kinase TaCIPK27 confers drought tolerance and exogenous ABA sensitivity in transgenic Arabidopsis. Plant Physiol Biochem 123, 103-113. |
| [88] | Weinl S, Kudla J (2009). The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184, 517-528. |
| [89] | Weng LY, Zhang MX, Wang K, Chen GL, Ding M, Yuan W, Zhu YY, Xu WF, Xu FY (2020). Potassium alleviates ammonium toxicity in rice by reducing its uptake through activation of plasma membrane H+-ATPase to enhance proton extrusion. Plant Physiol Biochem 151, 429-437. |
| [90] | Xi Y, Liu JY, Dong C, Cheng ZM (2017). The CBL and CIPK gene family in grapevine (Vitis vinifera): genome- wide analysis and expression profiles in response to various abiotic stresses. Front Plant Sci 8, 978. |
| [91] | Yang Y, Zhang C, Tang RJ, Xu HX, Lan WZ, Zhao FG, Luan F (2019). Calcineurin B-like proteins CBL4 and CBL10 mediate two independent salt tolerance pathways in Arabidopsis. Int J Mol Sci 20, 2421. |
| [92] | Yin X, Wang QL, Chen Q, Xiang N, Yang YQ, Yang YP (2017). Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase gene families in turnip ( Brassica rapa var. rapa). Front Plant Sci 8, 1191. |
| [93] | Yin XC, Xia YQ, Xie Q, Cao YX, Wang ZY, Hao GP, Song J, Zhou Y, Jiang XY (2020). The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance. J Exp Bot 71, 1801-1814. |
| [94] | Yu YH, Xia XL, Yin WL, Zhang HC (2007). Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul 52, 101-110. |
| [95] | Zhang HF, Yang B, Liu WZ, Li HW, Wang L, Wang YY, Deng M, Liang WW, Deyholos MK, Jiang YQ (2014a). Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol 14, 8. |
| [96] | Zhang HW, Feng H, Zhang JW, Ge RC, Zhang LY, Wang YX, Li LG, Wei JH, Li RF (2020). Emerging crosstalk between two signaling pathways coordinates K+ and Na+ homeostasis in the halophyte Hordeum brevisubulatum. J Exp Bot 71, 4345-4358. |
| [97] | Zhang T, Chen SX, Harmon AC (2014b). Protein phosphorylation in stomatal movement. Plant Signal Behav 9, e972845. |
| [98] | Zhang XX, Köster P, Schlücking K, Balcerowicz D, Hashimoto K, Kuchitsu K, Vissenberg K, Kudla J (2018b). CBL1-CIPK26-mediated phosphorylation enhances activity of the NADPH oxidase RBOHC, but is dispensable for root hair growth. FEBS Lett 592, 2582-2593. |
| [99] | Zhang Y, Lv Y, Jahan N, Chen G, Ren DY, Guo LB (2018a). Sensing of abiotic stress and ionic stress responses in plants. Int J Mol Sci 19, 3298. |
| [100] | Zhao JF, Yu AL, Du YW, Wang GH, Li YF, Zhao GY, Wang XD, Zhang WZ, Cheng K, Liu X, Wang ZH, Wang YW (2019). Foxtail millet (Setaria italica (L.) P. Beauv) CIPKs are responsive to ABA and abiotic stresses. PLoS One 14, e0225091. |
| [101] | Zheng XJ, He K, Kleist T, Chen F, Luan S (2015). Anion channel SLAH3 functions in nitrate-dependent alleviation of ammonium toxicity in Arabidopsis. Plant Cell Environ 38, 474-486. |
| [102] | Zhou XN, Hao HM, Zhang YG, Bai YL, Zhu WB, Qin YX, Yuan FF, Zhao FY, Wang MY, Hu JJ, Xu H, Guo AG, Zhao HX, Zhao Y, Cao CL, Yang YQ, Schumaker KS, Guo Y, Xie CG (2015). SOS2-like protein kinase 5, an SNF1-related protein kinase 3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of abscisic acid-insensitive 5. Plant Physiol 168, 659-676. |
| [103] | Zhou Y, Lai ZS, Yin XC, Yu S, Xu YY, Wang XX, Cong XL, Luo YH, Xu HX, Jiang XY (2016). Hyperactive mutant of a wheat plasma membrane Na+/H+ antiporter improves the growth and salt tolerance of transgenic tobacco. Plant Sci 253, 176-186. |
| [104] | Zhu JK (2016). Abiotic stress signaling and responses in plants. Cell 167, 313-324. |
| [105] | Zhu JK, Liu JP, Xiong LM (1998). Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell 10, 1181-1191. |
/
| 〈 |
|
〉 |