

Effects of Epigenetic Mechanisms on C4 Phosphoenolpyruvate Carboxylase Transgenic Rice (Oryza sativa) Seed Germination Under Drought Stress
Received date: 2020-03-21
Accepted date: 2020-08-26
Online published: 2020-09-03
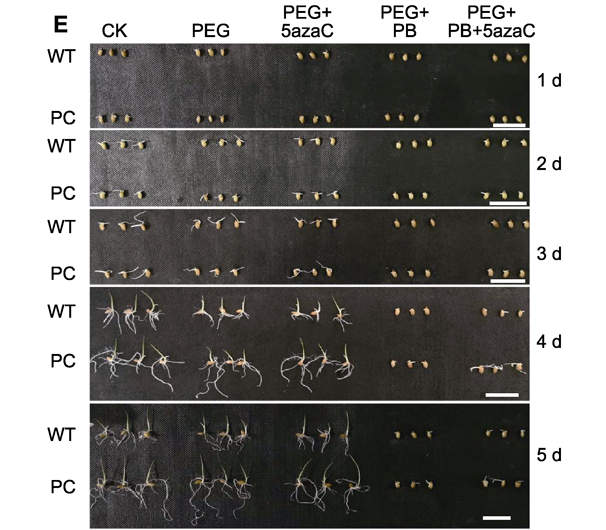
In order to reveal the effect of epigenetic mechanism under drought stress toward seed germination of transgenic rice within high maize C4-type PEPC gene expressing, C4-PEPC transgenic rice (PC) and wild type rice Kitaake (WT) were used in this study. By introducing DNA methylation inhibitor (5-azacytidine, 5azaC) and alternative splicing inhibitors (macrolides pladienolide B, PB), the drought simulation treatments with 10% (m/v) polyethylene glycol-6000 (PEG6000) alone or combining with the inhibitors were used for seed germination experiments. Seed vigor, soluble sugar and soluble protein content, α-amylase activity and the expression levels of related genes, PEPC-related genes, sugar signal-related genes, and some splicing factor genes during germination were measured. In the results, when treated with 0.25 µmol·L-1, PB had showed a significant inhibitory effect on the seed germination of the two tested rice lines under drought conditions. The content of total soluble sugar, sucrose, glucose, fructose and soluble protein during seed germination after PB addition was reduced to a certain extent under drought conditions. PB treatment also inhibited the gene expression of sucrose nonfermenting-1 (SNF1)-related protein kinase SnRKs family and the splicing factor arginine/serine-rich proteins (SR proteins), and the activity of α-amylase as well, but the inhibitory effect on PC is less than those on WT. 5 µmol·L -1 5azaC treatment had an opposite effect with alternative splicing inhibitors. The combination treatment with 5azaC and PEG6000 partially alleviated the inhibitory effect of drought stress on rice seed germination, and the germination rate of the tested materials increased. It can be seen that the mechanism of DNA methylation and alternative splicing are involved in drought tolerance at the bud stage of rice lines, with a larger effect on PC.
Ningxi Song, Yingfeng Xie, Xia Li . Effects of Epigenetic Mechanisms on C4 Phosphoenolpyruvate Carboxylase Transgenic Rice (Oryza sativa) Seed Germination Under Drought Stress[J]. Chinese Bulletin of Botany, 2020 , 55(6) : 677 -692 . DOI: 10.11983/CBB20048
| [1] | 陈蕾太, 孙爱清, 杨敏, 陈路路, 马雪丽, 李美玲, 尹燕枰 (2016). 基于小麦种子发芽逆境抗逆指数的种子活力评价. 应用生态学报 27, 2968-2974. |
| [2] | 杜康兮, 沈文辉, 董爱武 (2018). 表观遗传调控植物响应非生物胁迫的研究进展. 植物学报 53, 581-593. |
| [3] | 焦德茂, 匡廷云, 李霞, 戈巧英, 黄雪清, 郝乃斌, 白克智 (2003). 转PEPC基因水稻具有初级CO2浓缩机制的生理特点. 中国科学(C辑) 33, 33-39. |
| [4] | 焦德茂, 李霞, 黄雪清, 迟伟, 匡廷云, 古森本 (2001). 转PEPC基因水稻的光合CO2同化和叶绿素荧光特性. 科学通报 46, 414-418. |
| [5] | 李合生 (2000). 植物生理生化实验原理和技术 北京: 高等教育出版社. pp. 123-124. |
| [6] | 李美玲, 孙爱清, 杨敏, 张杰道, 王振林, 陈蕾太, 陈路路, 马雪丽, 尹燕枰 (2017). 小麦干热风抗性鉴定及热胁迫相关基因TaHSPs的表达分析. 麦类作物学报 37, 162-174. |
| [7] | 刘小龙, 李霞, 钱宝云 (2015). 外源Ca2+对PEG处理下转C4型PEPC基因水稻光合生理的调节 . 植物学报 50, 206-216. |
| [8] | 曲瑞莲, 吴春霞, 冯献忠 (2014). mRNA选择性剪切在植物发育中的作用. 植物生理学报 50, 717-724. |
| [9] | 宋凝曦, 张晓敬, 陆佳岚, 李霞, 谢寅峰 (2020). 可变剪接在植物响应胁迫中的作用. 植物生理学报 56, 1201-1211. |
| [10] | 张金飞, 李霞, 何亚飞, 谢寅峰 (2018). 外源葡萄糖增强高表达转玉米C4型PEPC水稻耐旱性的生理机制. 作物学报 44, 82-94. |
| [11] | 张金飞, 李霞, 谢寅峰 (2017). 植物SnRKs家族在胁迫信号通路中的调节作用. 植物学报 52, 346-357. |
| [12] | Ambavaram MMR, Basu S, Krishnan A, Ramegowda V, Batlang U, Rahman L, Baisakh N, Pereira A (2014). Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat Commun 5, 5302. |
| [13] | Bradford MM (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72, 248-254. |
| [14] | Brummell DA, Chen RKY, Harris JC, Zhang HB, Hamiaux C, Kralicek AV, McKenzie MJ (2011). Induction of vacuolar invertase inhibitor mRNA in potato tubers contributes to cold-induced sweetening resistance and includes spliced hybrid mRNA variants. J Exp Bot 62, 3519-3534. |
| [15] | Cao Y, Ma LG (2019). To splice or to transcribe: SKIP- mediated environmental fitness and development in plants. Front Plant Sci 10, 1222. |
| [16] | Chen PB, Li X, Huo K, Wei XD, Dai CC, Lv CG (2014). Promotion of photosynthesis in transgenic rice over-expressing of maize C4 phosphoenolpyruvate carboxylase gene by nitric oxide donors. J Plant Physiol 6, 458-466. |
| [17] | Cruz TMD, Carvalho RF, Richardson DN, Duque P (2014). Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression. Int J Mol Sci 15, 17541-17564. |
| [18] | Damaris RN, Lin ZY, Yang PF, He DL (2019). The rice α-amylase, conserved regulator of seed maturation and germination. Int J Mol Sci 20, 450. |
| [19] | Dinesh SP, Baker BJ (2000). Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc Nat Acad Sci USA 97, 1908-1913. |
| [20] | Dinkova TD, Márquez-Velázquez NA, Aguilar R, Lázaro- Mixteco PE, de Jiménez ES (2011). Tight translational control by the initiation factors eIF4E and eIF(iso)4E is required for maize seed germination. Seed Sci Res 21, 85-93. |
| [21] | Dong CL, He F, Berkowitz O, Liu JX, Cao PF, Tang M, Shi HC, Wang WJ, Li QL, Shen ZG, Whelan J, Zheng LQ (2018). Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice (Oryza sativa). Plant Cell 30, 2267-2285. |
| [22] | Egawa C, Kobayashi F, Ishibashi M, Nakamura T, Nakamura C, Takumi S (2006). Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet Syst 81, 77-91. |
| [23] | Filichkin SA, Cumbie JS, Dharmawardhana P, Jaiswal P, Chang JH, Palusa SG, Reddy ASN, Megraw M, Mockler TC (2015). Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol Plant 8, 207-227. |
| [24] | Foolad MR, Subbiah P, Zhang LP (2007). Common QTL affect the rate of tomato seed germination under different stress and nonstress conditions. Int J Plant Genomics 2007, 97386. |
| [25] | Gassmann W (2008). Alternative splicing in plant defense. In: Reddy ASN, Golovkin M, eds. Nuclear Pre-mRNA Processing in Plants. Berlin:Springer.pp. 219-233. |
| [26] | Golisz A, Sikorski PJ, Kruszka K, Kufel J (2013). Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res 41, 6232-6249. |
| [27] | Hakata M, Kuroda M, Miyashita T, Yamaguchi T, Kojima M, Sakakibara H, Mitsui T, Yamakawa H (2012). Suppression of α-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol J 10, 1110-1117. |
| [28] | He YF, Xie YF, Li X, Yang J (2020). Drought tolerance of transgenic rice overexpressing maize C4-PEPC gene related to increased anthocyanin synthesis regulated by sucrose and calcium. Biologia Plantarum 64, 136-149. |
| [29] | Hu QJ, Fu YY, Guan YJ, Lin C, Cao DD, Hu WM, Sheteiwy M, Hu J (2016). Inhibitory effect of chemical combinations on seed germination and pre-harvest sprouting in hybrid rice. Plant Growth Regul 80, 281-289. |
| [30] | Ku MSB, Agarie S, Nomura M, Fukayama H, Tsuchida H, Ono K, Hirose S, Toki S, Miyao M, Matsuoka M (1999). High-level expression of maize phosphoenolpyruvate carboxylase in transgenic rice plants. Nat Biotechnol 17, 76-80. |
| [31] | Lata C, Gupta S, Prasad M (2013). Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit Rev Biotechnol 33, 328-343. |
| [32] | Lebouteiller B, Gousset-Dupont A, Pierre JN, Bleton J, Tchapla A, Maucourt M, Moing A, Rolin D, Vidal J (2007). Physiological impacts of modulating phosphoenolpyruvate carboxylase levels in leaves and seeds of Arabidopsis thaliana. Plant Sci 172, 265-272. |
| [33] | Lev Maor G, Yearim A, Ast G (2015). The alternative role of DNA methylation in splicing regulation. Trends Genet 31, 274-280. |
| [34] | Li X, Wang C (2013). Physiological and metabolic enzymes activity changes in transgenic rice plants with increased phosphoenolpyruvate carboxylase activity during the flowering stage. Acta Physiol Plant 35, 1503-1512. |
| [35] | Li X, Wang C, Ren CG (2011). Effects of 1-butanol, neomycin and calcium on the photosynthetic characteristics of pepc transgenic rice. Afr J Biotechnol 10, 17466-17476. |
| [36] | Ling Y, Alshareef S, Butt H, Lozano-Juste J, Li LX, Galal AA, Moustafa A, Momin AA, Tashkandi M, Richardson DN, Fujii H, Arold S, Rodriguez PL, Duque P, Mahfouz MM (2017). Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J 89, 291-309. |
| [37] | Liu JJ, Sun N, Liu M, Liu JC, Du BJ, Wang XJ, Qi XT (2013). An autoregulatory loop controlling Arabidopsis HsfA2 expression: role of heat shock-induced alternative splicing. Plant Physiol 162, 512-521. |
| [38] | Liu XL, Li X, Zhang C, Dai CC, Zhou JY, Ren CG, Zhang JF (2017). Phosphoenolpyruvate carboxylase regulation in C4-PEPC-expressing transgenic rice during early responses to drought stress. Physiol Plant 159, 178-200. |
| [39] | Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using reatime quantitative PCR and the 2-ΔΔCt method . Methods 25, 402-408. |
| [40] | Marquez Y, Brown JWS, Simpson C, Barta A, Kalyna M (2012). Transcriptome survey reveals increased comp- lexity of the alternative splicing landscape in Arabidopsis. Genome Res 22, 1184-1195. |
| [41] | Marsh JT, Sullivan S, Hartwell J, Nimmo HG (2003). Structure and expression of phospho enolpyruvate carboxylase kinase genes in solanaceae. A novel gene exhibits alternative splicing. Plant Physiol 133, 2021-2028. |
| [42] | Miransari M, Smith DL (2014). Plant hormones and seed germination. Environ Exp Bot 99, 110-121. |
| [43] | Muthamilarasan M, Khandelwal R, Yadav CB, Bonthala VS, Khan Y, Prasad M (2014). Identification and molecular characterization of MYB transcription factor superfamily in C4 model plant foxtail millet ( Setaria italica L.). PLoS One 9, e109920. |
| [44] | Nakata M, Fukamatsu Y, Miyashita T, Hakata M, Kimura R, Nakata Y, Kuroda M, Yamaguchi T, Yamakawa H (2017). High temperature-induced expression of rice α-amylases in developing endosperm produces chalky grains. Front Plant Sci 8, 2089. |
| [45] | Nanjo Y, Asatsuma S, Itoh K, Hori H, Mitsui T (2004). Proteomic identification of α-amylase isoforms encoded by RAmy3B/3C from germinating rice seeds. Biosci Biotechnol Biochem 68, 112-118. |
| [46] | O’Leary B, Park J, Plaxton WC (2011). The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem J 436, 15-34. |
| [47] | Prasad PVV, Pisipati SR, Mom?ilovi? I, Ristic Z (2011). Independent and combined effects of high temperature and drought stress during grain filling on plant yield and chloroplast EF-Tu expression in spring wheat. J Agron Crop Sci 197, 430-441. |
| [48] | Qian BY, Li X, Liu XL, Wang M (2015). Improved oxidative tolerance in suspension-cultured cells of C4-pepc trans- genic rice by H2O2 and Ca2+ under PEG-6000. J Integr Plant Biol 57, 534-549. |
| [49] | Reddy ASN, Shad Ali G (2011). Plant serine/arginine-rich proteins: roles in precursor messenger RNA splicing, plant development, and stress responses. Wiley Interdiscip Rev RNA 2, 875-889. |
| [50] | Ren CG, Li X, Liu XL, Wei XD, Dai CC (2014). Hydrogen peroxide regulated photosynthesis in C4- pepc transgenic rice. Plant Physiol Biochem 74, 218-229. |
| [51] | Somani BL, Khanade J, Sinha R (1987). A modified anthrone-sulfuric acid method for the determination of fructose in the presence of certain proteins. Anal Biochem 167, 327-330. |
| [52] | Staiger D, Brown JWS (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25, 3640-3656. |
| [53] | Syed NH, Kalyna M, Marquez Y, Barta A, Brown JWS (2012). Alternative splicing in plants-coming of age. Trends Plant Sci 17, 616-623. |
| [54] | Tang YT, Li X, Lu W, Wei XD, Zhang QJ, Lv CG, Song NX (2018). Transgenic rice over-expressing maize C4 phosphoenolpyruvate carboxylase gene contributes to alleviating low nitrogen stress. Plant Physiol Biochem 130, 577-588. |
| [55] | Wang BB, Brendel V (2006). Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA 103, 7175-7180. |
| [56] | Yan HH, Kikuchi S, Neumann P, Zhang WL, Wu YF, Chen F, Jiang JM (2010). Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J 63, 353-365. |
| [57] | Zhang C, Li X, He YF, Zhang JF, Yan T, Liu XL (2017). Physiological investigation of C4-phosphoenolpyruvate- carboxylase-introduced rice line shows that sucrose metabolism is involved in the improved drought tolerance. Plant Physiol Biochem 115, 328-342. |
| [58] | Zhang P, Deng H, Xiao FM, Liu YS (2013). Alterations of alternative splicing patterns of Ser/Arg-rich (SR) genes in response to hormones and stresses treatments in different ecotypes of rice (Oryza sativa). J Integr Agric 12, 737-748. |
| [59] | Zhang XC, Gassmann W (2007). Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol 145, 1577-1587. |
/
| 〈 |
|
〉 |