

Received date: 2020-05-08
Accepted date: 2020-08-13
Online published: 2020-08-26
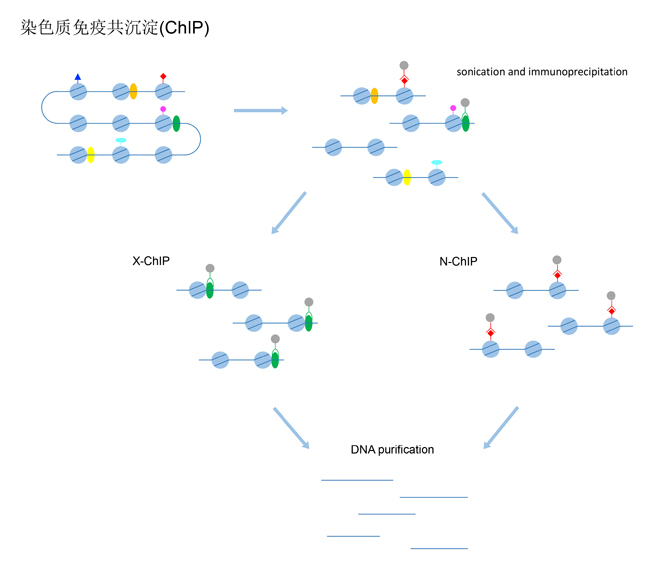
Chromatin immunoprecipitation (ChIP) is a teschnique used to investigate protein-DNA interaction. Briefly, protein and associated DNA are crosslinked, which is optional. Then the DNA-protein complexes are fragmented. By using an appropriate antibody, cross-linked DNA fragments associated with the protein of interest are selectively immunoprecipitated. ChIP is commonly used to identify binding regions of DNA-binding proteins, such as transcription factors. ChIP is also used to analyze histone modification profiles in combination with antibodies against specific histone modifications. Here we describe protocols and related tips for cross-linked ChIP and ultra-low-input micrococcal nuclease-based native ChIP (ULI-NChIP), which can be applied to rare cell populations at the 103 cell level.
Hongli Wang , Yuling Jiao . Protocols for Chromatin Immunoprecipitation[J]. Chinese Bulletin of Botany, 2020 , 55(4) : 475 -480 . DOI: 10.11983/CBB20076
| [1] | Adli M, Zhu J, Bernstein BE (2010). Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat Methods 7, 615-618. |
| [2] | Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823-837. |
| [3] | Brind’Amour J, Liu S, Hudson M, Chen C, Karimi MM, Lorincz MC (2015). An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun 6, 6033. |
| [4] | Collas P (2010). The current state of chromatin immunoprecipitation. Mol Biotechnol 45, 87-100. |
| [5] | Das PM, Ramachandran K, van Wert J, Singal R (2004). Chromatin immunoprecipitation assay. Biotechniques 37, 961-969. |
| [6] | Hitchler MJ, Rice JC (2011). Genome-wide epigenetic analysis of human pluripotent stem cells by ChIP and ChIP-Seq. In: Schwartz PH, Wesselschmidt RL, eds. Human Pluripotent Stem Cells: Methods and Protocols. New York: Humana Press. pp. 253-267. |
| [7] | Jackson V (1978). Studies on histone organization in the nucleosome using formaldehyde as a reversible cross- linking agent. Cell 15, 945-954. |
| [8] | Johnson KD, Bresnick EH (2002). Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods 26, 27-36. |
| [9] | Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig W, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553-560. |
| [10] | O'Neill LP, Turner BM (2003). Immunoprecipitation of native chromatin: NChIP. Methods 31, 76-82. |
| [11] | Orlando V (2000). Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci 25, 99-104. |
| [12] | Orlando V, Strutt H, Paro R (1997). Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11, 205-214. |
| [13] | Singal R, van Wert JM, Ferdinand L Jr (2002). Methylation of alpha-type embryonic globin gene alpha pi represses transcription in primary erythroid cells. Blood 100, 4217-4222. |
| [14] | Spencer VA, Sun JM, Li L, Davie JR (2003). Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods 31, 67-75. |
| [15] | Thorne AW, Myers FA, Hebbes TR (2004). Native chromatin immunoprecipitation. In: Tollefsbol TO, ed. Epigenetics Protocols. New York: Humana Press. pp. 21-44. |
| [16] | Wang J, Tian C, Zhang C, Shi B, Cao X, Zhang TQ, Zhao Z, Wang JW, Jiao Y (2017). Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 29, 1373-1387. |
| [17] | Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ (2001). Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol 21, 6820-6832. |
| [18] | Wells J, Farnham PJ (2002). Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods 26, 48-56. |
/
| 〈 |
|
〉 |