

Received date: 2020-05-28
Accepted date: 2020-06-02
Online published: 2020-06-11
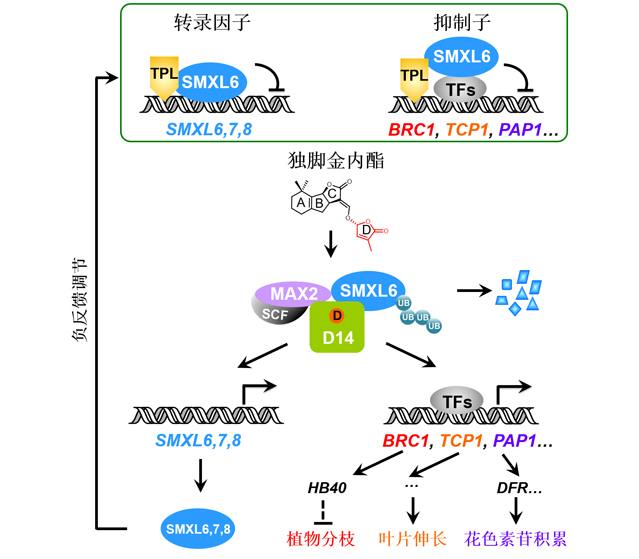
It is well-known that DELLA, AUX/IAA, JAZ and D53/SMXL act as repressor proteins that bind and repress transcription factors to suppress expression of hormone-responsive genes, while hormone molecules trigger signal transduction to induce degradation of these repressor proteins and eventually activate expression of hormone-responsive genes essential for various biological processes. The research team led by Dr. Jiayang Li recently reported that SMXL6, SMXL7 and SMXL8 (SMXL6,7,8) in strigolactone (SL) signaling pathway serve as dual-function repressor proteins which act as both repressors and transcription factors. They found that SMXL6,7,8 can function as transcription factors by directly binding to the promoters of SMXL6,7,8 genes and repressing their expression. In addition, they identified a large number of novel SL-responsive genes, and revealed molecular mechanisms underlying how SL regulates shoot branching, leaf elongation and anthocyanin biosynthesis. These important findings provide new insights into our understanding of plant hormone action, which are scientifically significant and agriculturally important.
Ruifeng Yao , Daoxin Xie . New Insight into Strigolactone Signaling[J]. Chinese Bulletin of Botany, 2020 , 55(4) : 397 -402 . DOI: 10.11983/CBB20099
| [1] | 黎家, 李传友 (2019). 新中国成立70年来植物激素研究进展. 中国科学: 生命科学 49, 1227-1281. |
| [2] | Bürger M, Chory J (2020). The many models of strigolactone signaling. Trends Plant Sci 25, 395-405. |
| [3] | de Saint Germain A, Clavé G, Badet-Denisot MA, Pillot JP, Cornu D, Le Caer JP, Burger M, Pelissier F, Retailleau P, Turnbull C, Bonhomme S, Chory J, Rameau C, Boyer FD (2016). A histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12, 787-794. |
| [4] | Duan J, Yu H, Yuan K, Liao Z, Meng X, Jing Y, Liu G, Chu J, Li J (2019). Strigolactone promotes cytokinin degradation through transcriptional activation of CYTOKININ OXIDASE/DEHYDROGENASE 9 in rice. Proc Natl Acad Sci USA 116, 14319-14324. |
| [5] | Fang Z, Ji Y, Hu J, Guo R, Sun S, Wang X (2020). Strigolactones and brassinosteroids antagonistically regulate the stability of the D53-OsBZR1 complex to determine FC1 expression in rice tillering. Mol Plant 13, 586-597. |
| [6] | Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, Bouwmeester H, Bécard G, Beveridge CA, Rameau C, Rochange SF (2008). Strigolactone inhibition of shoot branching. Nature 455, 189-194. |
| [7] | Hamiaux C, Drummond RSM, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012). DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22, 2032-2036. |
| [8] | Hu J, Ji Y, Hu X, Sun S, Wang X (2020). BES1 functions as the co-regulator of D53-like SMXLs to inhibit BRC1 expression in strigolactone-regulated shoot branching in Arabidopsis. Plant Commun 1, 100014. |
| [9] | Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, Yi W, Zhao L, Ma H, He Y, Wu Z, Melcher K, Qian Q, Xu H, Wang Y, Li J (2013). DWARF 53 acts as a repressor of strigolactone signaling in rice. Nature 504, 401-405. |
| [10] | Ma H, Duan J, Ke J, He Y, Gu X, Xu TH, Yu H, Wang Y, Brunzelle JS, Jiang Y, Rothbart SB, Xu H, Li J, Melcher K (2017). A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex. Sci Adv 3, e1601217. |
| [11] | Seto Y, Yasui R, Kameoka H, Tamiru M, Cao MM, Terauchi R, Sakurada A, Hirano R, Kisugi T, Hanada A, Umehara M, Seo E, Akiyama K, Burke J, Takeda-Kamiya N, Li WQ, Hirano Y, Hakoshima T, Mashiguchi K, Noel JP, Kyozuka J, Yamaguchi S (2019). Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun 10, 191. |
| [12] | Shabek N, Ticchiarelli F, Mao HB, Hinds TR, Leyser O, Zheng N (2018). Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signaling. Nature 563, 652-656. |
| [13] | Song X, Lu Z, Yu H, Shao G, Xiong J, Meng X, Jing Y, Liu G, Xiong G, Duan J, Yao X, Liu C, Li H, Wang Y, Li J (2017). IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res 27, 1128-1141. |
| [14] | Stanga JP, Smith SM, Briggs WR, Nelson DC (2013). SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol 163, 318-330. |
| [15] | Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J, Yamaguchi S (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195-200. |
| [16] | Uraguchi D, Kuwata K, Hijikata Y, Yamaguchi R, Imaizumi H, Am S, Rakers C, Mori N, Akiyama K, Irle S, McCourt P, Kinoshita T, Ooi T, Tsuchiya Y (2018). A femtomolar-range suicide germination stimulant for the parasitic plant Striga hermonthica. Science 362, 1301-1305. |
| [17] | Wang B, Wang Y, Li J (2017). Strigolactones. In: Li JY, Li CY, Smith SM, eds. Hormone Metabolism and Signaling in Plants. London: Academic Press. pp. 327-359. |
| [18] | Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J (2015). Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27, 3128-3142. |
| [19] | Wang L, Wang B, Yu H, Guo H, Lin T, Kou L, Wang A, Shao N, Ma H, Xiong G, Li X, Yang J, Chu J, Li J (2020a). Transcriptional regulation of strigolactone signaling in Arabidopsis. Nature 583, 277-281. |
| [20] | Wang L, Xu Q, Yu H, Ma H, Li X, Yang J, Chu J, Xie Q, Wang Y, Smith SM, Li J, Xiong G, Wang B (2020b). Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell 32, 2251-2270. |
| [21] | Waters MT, Gutjahr C, Bennett T, Nelson DC (2017). Strigolactone signaling and evolution. Annu Rev Plant Biol 68, 291-322. |
| [22] | Xie Y, Liu Y, Ma M, Zhou Q, Zhao Y, Zhao B, Wang B, Wei H, Wang H (2020). Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat Commun 11, 1955. |
| [23] | Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, Li Y, Yan C, Miao D, Sun Z, Yan J, Sun Y, Wang L, Chu J, Fan S, He W, Deng H, Nan F, Li J, Rao Z, Lou Z, Xie D (2016). DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469-473. |
| [24] | Yao R, Wang F, Ming Z, Du X, Chen L, Wang Y, Zhang W, Deng H, Xie D (2017). ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res 27, 838-841. |
| [25] | Yao R, Wang L, Li Y, Chen L, Li S, Du X, Wang B, Yan J, Li J, Xie D (2018). Rice DWARF14 acts as an unconventional hormone receptor for strigolactone. J Exp Bot 69, 2355-2365. |
| [26] | Zhao LH, Zhou XE, Wu ZS, Yi W, Xu Y, Li SL, Xu TH, Liu Y, Chen RZ, Kovach A, Kang YY, Hou L, He YZ, Xie C, Song WL, Zhong DF, Xu YC, Wang YH, Li JY, Zhang CH, Melcher K, Xu HE (2013). Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res 23, 436-439. |
| [27] | Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, Ma W, Gao H, Chen J, Yang C, Wang D, Tan J, Zhang X, Guo X, Wang J, Jiang L, Liu X, Chen W, Chu J, Yan C, Ueno K, Ito S, Asami T, Cheng Z, Wang J, Lei C, Zhai H, Wu C, Wang H, Zheng N, Wan J (2013). D14-SCFD3-dependent degradation of D53 regulates strigolactone signaling. Nature 504, 406-410. |
| [28] | Zwanenburg B, Blanco-Ania D (2018). Strigolactones: new plant hormones in the spotlight. J Exp Bot 69, 2205-2218. |
/
| 〈 |
|
〉 |