

The Development of Genomics Technologies Drives New Progress in Horticultural Plant Research
Received date: 2019-12-15
Accepted date: 2019-12-17
Online published: 2019-12-20
Horticultural plants include flowers, vegetables, fruit trees, some melon (such as watermelon (Citrullus lanatus), muskmelon (Cucumis melo)) and tea trees (Camellia sinensis), with a large number of species based on plant classification. Genomics and genetics researches for horticultural plants are of important theoretical value and economic significance. The development of genome sequencing technology and related bioinformatics tools greatly facilitate molecular biology researches of horticultural plants. In addition to its important ornamental value, the plant species ‘water lily’ has a very special position in evolutionary, belonging to an early angiosperm group. Recently, a high-quality genome map of an important flower plant, water lily, was generated. Through systematic analysis and genomic comparison of the water lily genome and other angiosperm genomes, the researchers thoroughly elucidated the evolutionary position and related evolutionary events of water lily. Based on the high-quality genomic sequences of these horticultural plants, researchers in horticultural plant science are expected to carry out in-depth molecular genetics research and identify functional genes underlying many traits such as flower organs, flower color, fragrance, and quality, which is expected to promote basic research and accelerate the creation of new varieties.
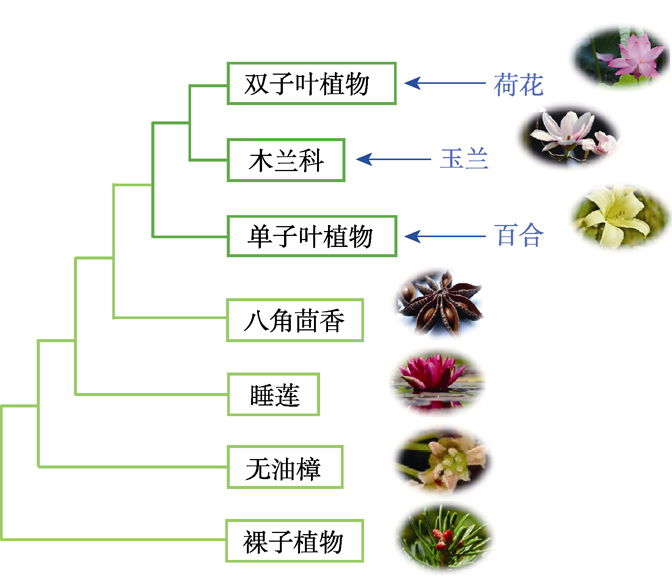
Key words: water lily; genome; horticulture; evolution
Jiali Tang , Jie Qiu , Xuehui Huang . The Development of Genomics Technologies Drives New Progress in Horticultural Plant Research[J]. Chinese Bulletin of Botany, 2020 , 55(1) : 1 -4 . DOI: 10.11983/CBB19240
| [1] | Chen F, Liu X, Yu CW, Chen YC, Tang HB, Zhang LS (2017). Water lilies as emerging models for Darwin’s abominable mystery. Hortic Res 4, 17051. |
| [2] | Chen LY, VanBuren R, Paris M, Zhou HY, Zhang XT, Wai CM, Yan HS, Chen S, Alonge M, Ramakrishnan S, Liao ZY, Liu J, Lin JS, Yue JJ, Fatima M, Lin ZC, Zhang JS, Huang LX, Wang H, Hwa TY, Kao SM, Choi AEY, Sharma A, Song J, Wang LL, Yim WC, Cushman JC, Paull RE, Matsumoto T, Qin Y, Wu QS, Wang JP, Yu QY, Wu J, Zhang SL, Boches P, Tung CW, Wang ML, D’Eeckenbrugge GC, Sanewski GM, Purugganan MD, Schatz MC, Bennetzen JL, Lexer C, Ming R (2019). The bracteatus pineapple genome and domestication of clonally propagated crops. Nat Genet 51, 1549-1558. |
| [3] | Guo SG, Zhao SJ, Sun HH, Wang X, Wu S, Lin T, Ren Y, Gao L, Deng Y, Zhang J, Lu XQ, Zhang HY, Shang JL, Gong GY, Wen CL, He N, Tian SW, Li MY, Liu JP, Wang YP, Zhu YC, Jarrets R, Levi A, Zhang XP, Huang SW, Fei ZJ, Liu WG, Xu Y (2019). Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat Genet 51, 1616-1623. |
| [4] | Liao NQ, Hu ZY, Li YY, Hao JF, Chen SN, Xue Q, Ma YY, Zhang KJ, Mahmoud A, Ali A, Malangisha GK, Lyu XL, Yang JH, Zhang MF (2020). Ethylene-responsive factor 4 is associated with the desirable rind hardness trait conferring cracking resistance in fresh fruits of watermelon. Plant Biol J 18, 1066-1077. |
| [5] | Ming R, VanBuren R, Wai CM, Tang HB, Schatz MC, Bowers JE, Lyons E, Wang ML, Chen J, Biggers E, Zhang JS, Huang LX, Zhang LM, Miao WJ, Zhang J, Ye ZY, Miao CY, Lin ZC, Wang H, Zhou HY, Yim WC, Priest HD, Zheng CF, Woodhouse M, Edger PP, Guyot R, Guo HB, Guo H, Zheng GY, Singh R, Sharma A, Min XJ, Zheng Y, Lee H, Gurtowski J, Sedlazeck FJ, Harkess A, McKain MR, Liao ZY, Fang JP, Liu J, Zhang XD, Zhang Q, Hu WC, Qin Y, Wang K, Chen LY, Shirley N, Lin YR, Liu LY, Hernandez AG, Wright CL, Bulone V, Tuskan GA, Heath K, Zee F, Moore PH, Sunkar R, Leebens-Mack JH, Mockler T, Bennetzen JL, Freeling M, Sankoff D, Paterson AH, Zhu XG, Yang XH, Smith JAC, Cushman JC, Paull RE, Yu QY (2015). The pineapple genome and the evolution of CAM photosynthesis. Nat Genet 47, 1435-1442. |
| [6] | The French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463-467. |
| [7] | Zhang LS, Chen F, Zhang XT, Li Z, Zhao YY, Rolf L, Chang XJ, Dong W, Simon YWH, Liu X, Song AX, Chen JH, Guo WL, Wang ZJ, Zhuang YY, Wang HF, Chen XQ, Hu J, Liu YH, Qin Y, Wang K, Dong SS, Liu Y, Zhang SZ, Yu XX, Wu Q, Wang LS, Yan XQ, Jiao YN, Kong HZ, Zhou XF, Yu CW, Chen YC, Li F, Wang JH, Chen W, Chen XL, Jia QD, Zhang C, Jiang YF, Zhang WB, Liu GH, Fu JY, Chen F, Ma H, Yves VP, Tang HB (2019). The water lily genome and the early evolution of flowering plants. Nature 577, 79-84. |
| [8] | Zhao GW, Lian Q, Zhang ZH, Fu QS, He YH, Ma S, Ruggieri V, Monforte AJ, Wang PY, Julca I, Wang HS, Liu JP, Xu Y, Wang RZ, Ji JB, Xu ZH, Kong WH, Zhong Y, Shang JL, Pereira L, Argyris J, Zhang J, Mayobre C, Pujol M, Oren E, Out D, Wang JM, Sun DX, Zhao SJ, Zhu YC, Li N, Katzir N, Gur A, Dogimont C, Schaefer H, Fan W, Bendahmane A, Fei Z, Pitrat M, Gabaldon T, Lin T, Garcia-Mas J, Xu YY, Huang SW (2019). A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat Genet 51, 1607-1615. |
/
| 〈 |
|
〉 |