

Stress Memory Mediated by Epigenetic Modification in Plants
Received date: 2019-07-18
Accepted date: 2019-10-17
Online published: 2019-10-18
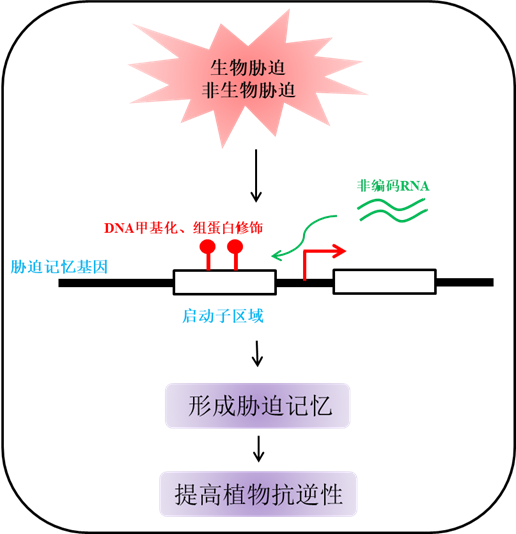
Because of the fixed growth habits lacking of mobility, plants have innovated unique strategies to cope with variable environmental conditions. For their survival, plants have evolved mechanisms of stress memories to adapt to the adverse environments and thus protect themselves. Epigenetic modifications not only regulate the growth and development of plants, but also participate in responses to various abiotic and/or biotic stresses. Recent studies have shown that epigenetic modifications play important roles in the control of plant stress memory. In particular, DNA methylation, histone methylation, histone acetylation modification, and other modifications are involved in the formation and the maintenance of specific stress memories. This review highlights the recent advances of plant stress memories mediated by epigenetic modifications, and some key challenges in this field were discussed.
Wei Chen , Yingzeng Yang , Feng Chen , Wenguan Zhou , Kai Shu . Stress Memory Mediated by Epigenetic Modification in Plants[J]. Chinese Bulletin of Botany, 2019 , 54(6) : 779 -785 . DOI: 10.11983/CBB19137
| [1] | Bannister AJ, Schneider R, Kouzarides T ( 2002). Histone methylation: dynamic or static? Cell 109, 801-806. |
| [2] | Berry S, Dean C ( 2015). Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J 83, 133-148. |
| [3] | Bouché F, Woods DP, Amasino RM ( 2017). Winter memory throughout the plant kingdom: different paths to flowering. Plant Physiol 173, 27-35. |
| [4] | Boyko A, Blevins T, Yao YL, Golubov A, Bilichak A, Ilnytskyy Y, Hollander J, Meins F Jr, Kovalchuk I ( 2010). Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of dicer-like proteins. PLoS One 5, e9514. |
| [5] | Brzezinka K, Altmann S, Czesnick H, Nicolas P, Gorka M, Benke E, Kabelitz T, J?hne F, Graf A, Kappel C, B?urle I ( 2016). Arabidopsis FORGETTER1 mediates stress- induced chromatin memory through nucleosome remodeling. eLife 5, e17061. |
| [6] | Chong SY, Whitelaw E ( 2004). Epigenetic germline inheritance. Curr Opin Genet Dev 14, 692-696. |
| [7] | Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR ( 2015). Priming for enhanced defense. Annu Rev Phytopathol 53, 97-119. |
| [8] | Ding Y, Fromm M, Avramova Z ( 2012). Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat Commun 3, 740. |
| [9] | Ding Y, Liu N, Virlouvet L, Riethoven JJ, Fromm M, Avramova Z ( 2013). Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol 13, 229. |
| [10] | Ding Y, Virlouvet L, Liu N, Riethoven JJ, Fromm M, Avramova Z ( 2014). Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biol 14, 141. |
| [11] | Eberharter A, Becker PB ( 2002). Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep 3, 224-229. |
| [12] | Ebrahim A, Brunk E, Tan J, O’Brien EJ, Kim D, Szubin R, Lerman JA, Lechner A, Sastry A, Bordbar A, Feist AM, Palsson BO ( 2016). Multi-omic data integration enables discovery of hidden biological regularities. Nat Commun 7, 13091. |
| [13] | Feng QZ, Yang CW, Lin XY, Wang JM, Ou XF, Zhang CY, Chen Y, Liu B ( 2012). Salt and alkaline stress induced transgenerational alteration in DNA methylation of rice ( Oryza sativa ). Aust J Crop Sci 6, 877-883. |
| [14] | Feng XJ, Li JR, Qi SL, Lin QF, Jin JB, Hua XJ ( 2016). Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc Natl Acad Sci USA 113, E8335-E8343. |
| [15] | Franks SJ, Hoffmann AA ( 2012). Genetics of climate change adaptation. Annu Rev Genet 46, 185-208. |
| [16] | Friedrich T, Faivre L, B?urle I, Schubert D ( 2019). Chromatin-based mechanisms of temperature memory in plants. Plant Cell Environ 42, 762-770. |
| [17] | Gruenbaum Y, Naveh-Many T, Cedar H, Razin A ( 1981). Sequence specificity of methylation in higher plant DNA. Nature 292, 860-862. |
| [18] | Herman JJ, Sultan SE ( 2016). DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Proc R Soc B 283, 20160988. |
| [19] | Hilker M, Schmülling T ( 2019). Stress priming, memory, and signaling in plants. Plant Cell Environ 42, 753-761. |
| [20] | Jaskiewicz M, Conrath U, Peterh?nsel C ( 2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12, 50-55. |
| [21] | Kwon CS, Lee D, Choi G, Chung WI ( 2009). Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J 60, 112-121. |
| [22] | L?mke J, B?urle I ( 2017). Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol 18, 124. |
| [23] | L?mke J, Brzezinka K, Altmann S, B?urle I ( 2016). A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J 35, 162-175. |
| [24] | Li P, Yang H, Wang L, Liu HJ, Huo HQ, Zhang CJ, Liu AZ, Zhu AD, Hu JY, Lin YJ, Liu L ( 2019). Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front Genet 10, 55. |
| [25] | Liu HC, L?mke J, Lin SY, Hung MJ, Liu KM, Charng YY, B?urle I ( 2018). Distinct heat shock factors and chromatin modifications mediate the organ-autonomous transcriptional memory of heat stress. Plant J 95, 401-413. |
| [26] | Liu JZ, Feng LL, Gu XT, Deng X, Qiu Q, Li Q, Zhang YY, Wang MY, Deng YW, Wang ET, He YK, B?urle I, Li JM, Cao XF, He ZH ( 2019). An H3K27me3 demethylase- HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res 29, 379-390. |
| [27] | Luna E, Bruce TJA, Roberts MR, Flors V, Ton J ( 2012). Next-generation systemic acquired resistance. Plant Physiol 158, 844-853. |
| [28] | Martinez-Medina A, Flors V, Heil M, Mauch-Mani B, Pieterse CMJ, Pozo MJ, Ton J, van Dam NM, Conrath U ( 2016). Recognizing plant defense priming. Trends Plant Sci 21, 818-822. |
| [29] | Molinier J, Ries J, Zipfel C, Hohn B ( 2006). Transgeneration memory of stress in plants. Nature 442, 1046-1049. |
| [30] | Rasmann S, de Vos M, Casteel CL, Tian DL, Halitschke R, Sun JY, Agrawal AA, Felton GW, Jander G ( 2012). Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol 158, 854-863. |
| [31] | Richards EJ ( 2006). Inherited epigenetic variation-revisiting soft inheritance. Nat Rev Genet 7, 395-401. |
| [32] | Sani E, Herzyk P, Perrella G, Colot V, Amtmann A ( 2013). Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol 14, R59. |
| [33] | Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, B?urle I ( 2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792-1807. |
| [34] | van Hulten M, Pelser M, van Loon LC, Pieterse CMJ, Ton J ( 2006). Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA 103, 5602-5607. |
| [35] | Vanyushin BF, Ashapkin VV ( 2011). DNA methylation in higher plants: past, present and future. Biochim Biophys Acta Gene Regul Mech 1809, 360-368. |
| [36] | Wang JJ, Meng XW, Dobrovolskaya OB, Orlov YL, Chen M ( 2017). Non-coding RNAs and their roles in stress response in plants. Genom Proteom Bioinformat 15, 301-312. |
| [37] | Wibowo A, Becker C, Marconi G, Durr J, Price J, Hagmann J, Papareddy R, Putra H, Kageyama J, Becker J, Weigel D, Gutierrez-Marcos J ( 2016). Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 5, e13546. |
| [38] | Yakovlev IA, Carneros E, Lee YK, Olsen JE, Fossdal CG ( 2016). Transcriptional profiling of epigenetic regulators in somatic embryos during temperature induced formation of an epigenetic memory in Norway spruce. Planta 243, 1237-1249. |
| [39] | Zhang XY, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE ( 2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol 10, R62. |
/
| 〈 |
|
〉 |