

An Quick and Efficient Assay for In Vivo Protein Ubiquitination
Received date: 2019-08-12
Accepted date: 2019-09-24
Online published: 2019-10-09
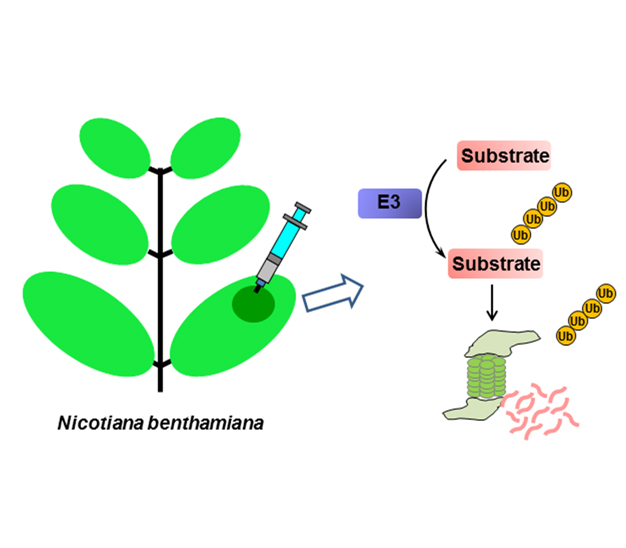
The UPS (ubiquitination/proteasome system) plays a vital role in nearly all plant signaling processes, for example, jasmonic acid receptor COI1 (coronotine insensitive protein 1) and auxin receptor TIR1 (transport inhibitor response 1) are F-box type E3s ligases, and they promote the ubiquitination then degradation of specific transcriptional repressors through 26S proteasome to activate hormone signaling. However, for the whole UPS system, only a few E3 ligase/substrate pairs’ interactions have been demonstrated due to technical limitations in plants. Generally, E3 ligase and substrate are expressed in Escherichia coli for ubiquitination assay, which may lack post-translational modifications that are important for protein function, then gives false negative result. We describe a quick and efficient assay for detecting E3-mediated protein ubiquitination in vivo by means of agroinfiltration for transient expression of relevant genes in tobacco (Nicotiana benthamiana) leaves. In detail, this method can detect the specific interaction between E3 ligase and substrate, substrate ubiquitination, the effect of E3 ligase on degradation of its substrate, inhibition of substrate degradation by 26S proteasome inhibitor MG132, and in vitro ubiquitination assay with endogenous plant proteins.
Lijing Liu , Qingzhen Zhao , Qi Xie , Feifei Yu . An Quick and Efficient Assay for In Vivo Protein Ubiquitination[J]. Chinese Bulletin of Botany, 2019 , 54(6) : 753 -763 . DOI: 10.11983/CBB19153
| [1] | 赵庆臻, 刘利静, 谢旗, 于菲菲 ( 2019). 植物蛋白的体外泛素化检测方法. 植物学报 54, 764-772. |
| [2] | Chakrabarty R, Banerjee R, Chung SM, Farman M, Citovsky V, Hogenhout SA, Tzfira T, Goodin M ( 2007). PSITE vectors for stable integration or transient expression of autofluorescent protein fusions in plants: probing Nicotiana benthamiana -virus interactions. Mol Plant Microbe Interact 20, 740-750. |
| [3] | Chen G, Huang H, Frohlich O, Yang Y, Klein JD, Price SR, Sands JM ( 2008). MDM2 E3 ubiquitin ligase mediates UT-A1 urea transporter ubiquitination and degradation. Am J Physiol Renal Physiol 295, F1528-F1534. |
| [4] | Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM ( 2000). MDM2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275, 8945-8951. |
| [5] | Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO ( 2002). pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31, 375-383. |
| [6] | Johansen LK, Carrington JC ( 2001). Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium -mediated transient expression system. Plant Physiol 126, 930-938. |
| [7] | Ko?ciańska E, Kalantidis K, Wypijewski K, Sadowski J, Tabler M ( 2005). Analysis of RNA silencing in agroinfiltrated leaves of Nicotiana benthamiana and Nicotiana tabacum. Plant Mol Biol 59, 647-661. |
| [8] | Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT ( 2009). Drought stress-induced Rma1H1, a RING membrane- anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21, 622-641. |
| [9] | Liu H, Stone SL ( 2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22, 2630-2641. |
| [10] | Liu LL, Zhang YY, Tang SY, Zhao QZ, Zhang ZH, Zhang HW, Dong L, Guo HS, Xie Q ( 2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61, 893-903. |
| [11] | Ma P, Liu J, He H, Yang M, Li M, Zhu X, Wang X ( 2009). A viral suppressor P1/HC-pro increases the GFP gene expression in Agrobacterium-mediated transient assay. Appl Biochem Biotechnol 158, 243-252. |
| [12] | Mokrzycki-Issartel N, Bouchon B, Farrer S, Berland P, Laparra H, Madelmont JC, Theisen M ( 2003). A transient tobacco expression system coupled to MALDI-TOFMS allows validation of the impact of differential targeting on structure and activity of a recombinant therapeutic glycoprotein produced in plants. FEBS Lett 552, 170-176. |
| [13] | Osterlund MT, Hardtke CS, Wei N, Deng XW ( 2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462-466. |
| [14] | Proietto M, Bianchi MM, Ballario P, Brenna A ( 2015). Epigenetic and posttranslational modifications in light signal transduction and the circadian clock in Neurospora crassa. Int J Mol Sci 16, 15347-15383. |
| [15] | Rodriguez M, Ramírez NI, Ayala M, Freyre F, Pérez L, Triguero A, Mateo C, Selman-Housein G, Gavilondo JV, Pujol M ( 2005). Transient expression in tobacco leaves of an aglycosylated recombinant antibody against the epidermal growth factor receptor. Biotechnol Bioeng 89, 188-194. |
| [16] | Smalle J, Vierstra RD ( 2004). The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55, 555-590. |
| [17] | Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J ( 2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415-3428. |
| [18] | Ueki S, Lacroix B, Krichevsky A, Lazarowitz SG, Citovsky V ( 2009). Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat Protoc 4, 71-77. |
| [19] | Vierstra RD ( 2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci 8, 135-142. |
| [20] | Vierstra RD ( 2012). The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol 160, 2-14. |
| [21] | Voinnet O, Rivas S, Mestre P, Baulcombe D ( 2003). An enhanced transient expression system in plants based on suppression of gene silencing by the P19 protein of tomato bushy stunt virus. Plant J 33, 949-956. |
| [22] | Wroblewski T, Tomczak A, Michelmore R ( 2005). Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol J 3, 259-273. |
| [23] | Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH ( 2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167-170. |
| [24] | Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, Zhang Y, Lee I, Xie Q, Paek NC, Deng XW ( 2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32, 617-630. |
/
| 〈 |
|
〉 |