

NRT1.1B Connects Root Microbiota and Nitrogen Use in Rice
Received date: 2019-03-31
Accepted date: 2019-04-02
Online published: 2019-04-29
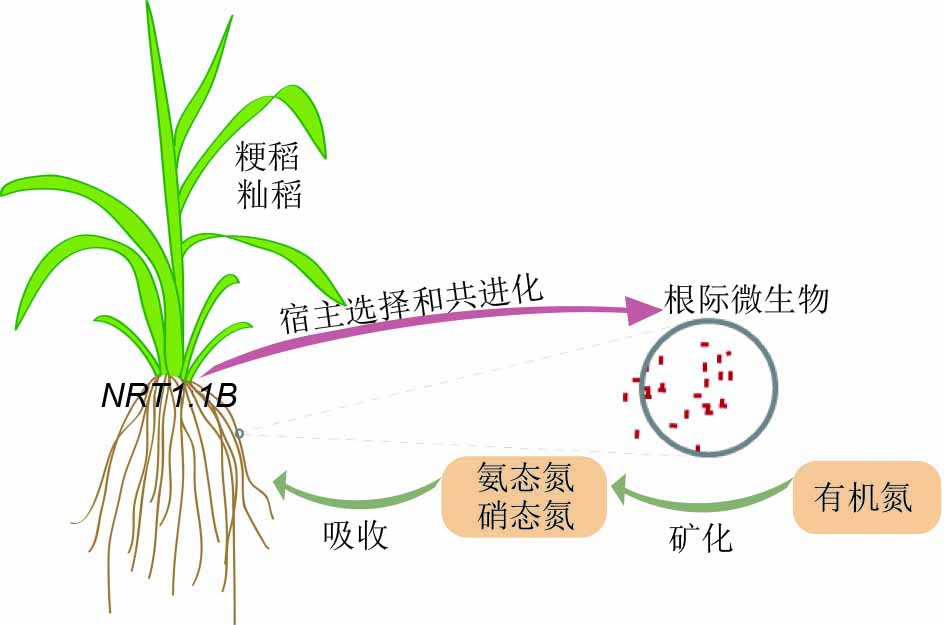
Root-associated microbial communities in the soil play fundamental roles in plant nutrition uptake and fitness. However, how plants shape root microbial communities and how the microbes affect the fitness of their hosts remain elusive. Recently, Chinese scientists have made a breakthrough discovery that the nitrogen-use efficiency between indica and japonica rice varieties is associated with different root microbiota in rice. Nitrogen metabolism is greatly enriched in indica-enriched bacteria as compared with japonica-enriched bacteria. Rice NRT1.1B, a nitrogen sensor contributing to nitrogen use divergence between rice subspecies, is associated with the recruitment of these bacterial taxa. Inoculation of the japonica variety with indica-enriched bacteria can improve rice growth in organic nitrogen conditions in the SynCom experimental system. This work highlights the links between root microbiota and nitrogen use in rice and could be exploited to modulate the root microbiota that increase crop productivity and sustainability.
Key words: rice; NRT1.1B; root microbiota; nitrogen use
Xiaolin Wang , Ertao Wang . NRT1.1B Connects Root Microbiota and Nitrogen Use in Rice[J]. Chinese Bulletin of Botany, 2019 , 54(3) : 285 -287 . DOI: 10.11983/CBB19060
| [1] | Beckers B, Op De Beeck M, Weyens N, Van Acker R, Van Montagu M, Boerjan W, Vangronsveld J ( 2016). Lignin engineering in field-grown poplar trees affects the endosphere bacterial microbiome. Proc Natl Acad Sci USA 113, 2312-2317. |
| [2] | Bender SF, Wagg C, van der Heijden MGA ( 2016). An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol 31, 440-452. |
| [3] | Berendsen RL, Pieterse CM, Bakker PA ( 2012). The rhizosphere microbiome and plant health. Trends Plant Sci 17, 478-486. |
| [4] | Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P ( 2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91-95. |
| [5] | Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P ( 2013). Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64, 807-838. |
| [6] | Chaparro JM, Sheflin AM, Manter DK, Vivanco JM ( 2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fert Soils 48, 489-499. |
| [7] | Duan L, Liu HB, Li XH, Xiao JH, Wang SP ( 2014). Multiple phytohormones and phytoalexins are involved in disease resistance to Magnaporthe oryzae invaded from roots in rice. Physiol Plant 152, 486-500. |
| [8] | Edwards J, Johnson C, Santos-Medellin C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V ( 2015). Structure, variation, and assembly of the root- associated microbiomes of rice. Proc Natl Acad Sci USA 112, E911-E920. |
| [9] | Hu B, Wang W, Ou SJ, Tang JY, Li H, Che RH, Zhang ZH, Chai XY, Wang HR, Wang YQ, Liang CZ, Liu LC, Piao ZZ, Deng QY, Deng K, Xu C, Liang Y, Zhang LH, Li LG, Chu CC ( 2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47, 834-838. |
| [10] | Mbodj D, Effa-Effa B, Kane A, Manneh B, Gantet P, Laplaze L, Diedhiou AG, Grondin A ( 2018). Arbuscular mycorrhizal symbiosis in rice: establishment, environmental control and impact on plant growth and resistance to abiotic stresses. Rhizosphere 8, 12-26. |
| [11] | Oldroyd GED, Murray JD, Poole PS, Downie JA ( 2011). The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45, 119-144. |
| [12] | Santhanam R, Luu VT, Weinhold A, Goldberg J, Oh Y, Baldwin IT ( 2015). Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc Natl Acad Sci USA 112, E5013-E5020. |
| [13] | Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, Fukuda S, Ushio M, Nakaoka S, Onoda Y, Yoshida K, Schlaeppi K, Bai Y, Sugiura R, Ichihashi Y, Minamisawa K, Kiers ET ( 2018). Core microbiomes for sustainable agroecosystems. Nat Plants 4, 247-257. |
| [14] | Walters WA, Jin Z, Youngblut N, Wallace JG, Sutter J, Zhang W, Gonzalez-Pena A, Peiffer J, Koren O, Shi Q, Knight R, Glavina del Rio T, Tringe SG, Buckler ES, Dangl JL, Ley RE ( 2018). Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc Natl Acad Sci USA 115, 7368-7373. |
| [15] | Zhang J, Liu Y, Zhang N, Hu B, Jin T, Xu H, Qin Y, Yan P, Zhang X, Guo X, Hui J, Cao S, Wang X, Wang C, Wang H, Qu B, Fan GY, Yuan L, Garrido-Oter R, Chu C, Bai Y ( 2019). NRT1.1B contributes the association of root microbiota and nitrogen use in rice. Nat Biotechnol doi: https://doi.org/10.1038/s41587-019-0104-4. |
/
| 〈 |
|
〉 |