

High-throughput Identification of Meiotic Anti-CO Mutants by Fluorescent Reporters
Received date: 2018-10-30
Accepted date: 2019-02-11
Online published: 2019-02-21
The forward genetic approach has become a widespread methodology to reveal genetic factors involved in meiosis, such as the crossover negative regulators, which limit the class II crossover formation. Here we developed a forward genetics mutant screen to identify anti-CO mutants under the Col wild-type background of Arabidopsis thaliana. We isolated 18 mutant lines showing more than three-fold increase in male meiotic recombination frequency as compared with the wild type, including dominant and recessive mutants. Thus, the EMS screen based on fluorescent reporters allows for high-throughput identification of meiotic anti-CO mutants and provides a novel approach and genetic materials to study the molecular mechanism of meiotic recombination regulation.
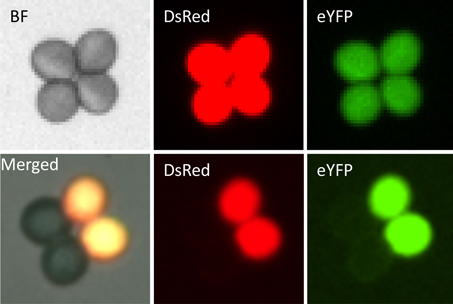
Key words: meiosis; anti-CO mutants; fluorescent marker; high-throughput visual assay
Fan Li , Jiwei Ruan . High-throughput Identification of Meiotic Anti-CO Mutants by Fluorescent Reporters[J]. Chinese Bulletin of Botany, 2019 , 54(4) : 522 -530 . DOI: 10.11983/CBB18229
| [1] | Berchowitz LE, Copenhaver GP ( 2008). Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc 3, 41-50. |
| [2] | Berchowitz LE, Copenhaver GP ( 2010). Genetic interference: don't stand so close to me. Curr Genomics 11, 91-102. |
| [3] | Berchowitz LE, Francis KE, Bey AL, Copenhaver GP ( 2007). The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet 3, e132. |
| [4] | Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R ( 2012). FANCM limits meiotic crossovers. Science 336, 1588-1590. |
| [5] | Fernandes JB, Duhamel M, Seguéla-Arnaud M, Froger N, Girard C, Choinard S, Solier V, De Winne N, De Jaeger G, Gevaert K, Andrey P, Grelon M, Guerois R, Kumar R, Mercier R ( 2017). FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. PLoS Genet 14, e1007317. |
| [6] | Fernandes JB, Séguéla-Arnaud M, Larchevêque C, Lloyd AH, Mercier R ( 2018). Unleashing meiotic crossovers in hybrid plants. Proc Natl Acad Sci USA 115, 2431-2436. |
| [7] | Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP ( 2007). Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci USA 104, 3913-3918. |
| [8] | Girard C, Chelysheva L, Choinard S, Froger N, Macaisne N, Lehmemdi A, Mazel J, Crismani W, Mercier R ( 2015). AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet 11, e1005369. |
| [9] | Girard C, Crismani W, Froger N, Mazel J, Lemhemdi A, Horlow C, Mercier R ( 2014). FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res 42, 9087-9095. |
| [10] | Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C ( 2011). Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7, e1002354. |
| [11] | Hatkevich T, Kohl KP, McMahan S, Hartmann MA, Williams AM, Sekelsky J ( 2017). Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr Biol 27, 96-102. |
| [12] | Higgins JD, Buckling EF, Franklin FC, Jones GH ( 2008). Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54, 152-162. |
| [13] | Hu Q, Li YF, Wang HJ, Shen Y, Zhang C, Du GJ, Tang D, Cheng ZK ( 2017). Meiotic chromosome association 1 interacts with TOP3α and regulates meiotic recombination in rice. Plant Cell 29, 1697-1708. |
| [14] | Huang JY, Cheng ZH, Wang C, Hong Y, Su H, Wang J, Copenhaver GP, Ma H, Wang YX ( 2015). Formation of interference-sensitive meiotic cross-overs requires sufficient DNA leading-strand elongation. Proc Natl Acad Sci USA 112, 12534-12539. |
| [15] | Jones GH, Franklin FCH ( 2006). Meiotic crossing-over: obligation and interference. Cell 126, 246-248. |
| [16] | Kim Y, Schumaker KS, Zhu JK ( 2006). EMS mutagenesis of Arabidopsis. In: Salinas J, Sanchez-Serrano JJ, eds. Arabidopsis Protocols. Totowa: Humana Press. pp. 101-103. |
| [17] | Kurzbauer MT, Pradillo M, Kerzendorfer C, Sims J, Ladurner R, Oliver C, Janisiw MP, Mosiolek M, Schweizer D, Copenhaver GP, Schl?gelhofer P ( 2018). Arabidopsis thaliana FANCD2 promotes meiotic crossover formation. Plant Cell 30, 415-428. |
| [18] | Li F, De Storme N, Geelen D ( 2017). Dynamics of male meiotic recombination frequency during plant development using fluorescent tagged lines in Arabidopsis thaliana. Sci Rep 7, 42535. |
| [19] | Lu PL, Han XW, Qi J, Yang JG, Wijeratne AJ, Li T, Ma H ( 2012). Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res 22, 508-518. |
| [20] | Lu PL, Wijeratne AJ, Wang ZJ, Copenhaver GP, Ma H ( 2014). Arabidopsis PTD is required for type I crossover formation and affects recombination frequency in two different chromosomal regions. J Genet Genomics 41, 165-175. |
| [21] | Lukowitz W, Gillmor CS, Scheible WR ( 2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123, 795-806. |
| [22] | Macaisne N, Novatchkova M, Peirera L, Vezon D, Jolivet S, Froger N, Chelysheva L, Grelon M, Mercier R ( 2008). SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr Biol 18, 1432-1437. |
| [23] | Macaisne N, Vignard J, Mercier R ( 2011). SHOC1 and PTD form an XPF-ERCC1-like complex that is required for formation of class I crossovers. J Cell Sci 124, 2687-2691. |
| [24] | Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M ( 2015). The molecular biology of meiosis in plants. Annu Rev Plant Biol 66, 297-327. |
| [25] | Mieulet D, Aubert G, Bres C, Klein A, Droc G, Vieille E, Rond-Coissieux C, Sanchez M, Dalmais M, Mauxion JP, Rothan C, Guiderdoni E, Mercier R ( 2018). Unleashing meiotic crossovers in crops. Nat Plants 4, 1010-1016. |
| [26] | Qi J, Chen YM, Copenhaver GP, Ma H ( 2014). Detection of genomic variations and DNA polymorphisms and impact on analysis of meiotic recombination and genetic mapping. Proc Natl Acad Sci USA 111, 10007-10012. |
| [27] | Qu LJ, Qin GJ ( 2014). Generation and identification of Arabidopsis EMS mutants. In: Sanchez-Serrano JJ, Salinas J, eds. Arabidopsis Protocols. Totowa: Humana Press. pp. 225-239. |
| [28] | Séguéla-Arnaud M, Choinard S, Larchevêque C, Girard C, Froger N, Crismani W, Mercier R ( 2017). RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res 4, 1860-1871. |
| [29] | Séguéla-Arnaud M, Crismani W, Larchevêque C, Mazel J, Froger N, Choinard S, Lemhemdi A, Macaisne N, Van Leene J, Gevaert K, De Jaeger G, Chelysheva L, Mercier R ( 2015). Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc Natl Acad Sci USA 112, 4713-4718. |
| [30] | Wang YX, Cheng ZH, Huang JY, Shi Q, Hong Y, Copenhaver GP, Gong ZZ, Ma H ( 2012). The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet 8, e1003039. |
| [31] | Yelina NE, Lambing C, Hardcastle TJ, Zhao XH, Santos B, Henderson IR ( 2015). DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Gene Dev 29, 2183-2202. |
| [32] | Yelina NE, Ziolkowski PA, Miller N, Zhao X, Kelly KA, Mu?oz DF, Mann DJ, Copenhaver GP, Henderson IR ( 2013). High-throughput analysis of meiotic crossover frequency and interference via flow cytometry of fluorescent pollen in Arabidopsis thaliana. Nat Protoc 8, 2119-2134. |
/
| 〈 |
|
〉 |