

EARLY SENESCENCE 1 Participates in the Expression Regulation of Circadian Clock Genes and Response to Stress in Rice
Received date: 2016-01-26
Revised date: 2016-05-28
Online published: 2016-12-02
Plant circadian clock genes help control the activities of all growth stages and development. We cloned EARLY SENESCENCE 1 (ES1), which affects water loss in rice, and used a segregation mutant es1-1 and the wild type (NPB) for microarray assay of the root and leaf tissue. The mutant es1-1 had 42 major upregulated genes and 14 major downregulated genes involved in 24 metabolic pathways, including regulating the circadian clock (n=4), methane metabolism (n=3), and phenylalanine metabolism (n=3). On expression profiling of rice circadian clock-related genes, the expression in es1-1 differed from that in the wild type. After cold treatment, es1-1 exhibited more cold tolerance than the wild type, and expression analysis of the circadian clock-related genes revealed changes in es1-1 and NPB. Moreover, after inoculating Xanthomonas oryzae pv. oryzae, es1-1 showed enhanced resistance during tillering as compared with the wild type. ES1 may participate in the expression regulation of rice circadian clock genes, as well as response to some certain stress, which provide further clues for research of rice circadian clock genes.
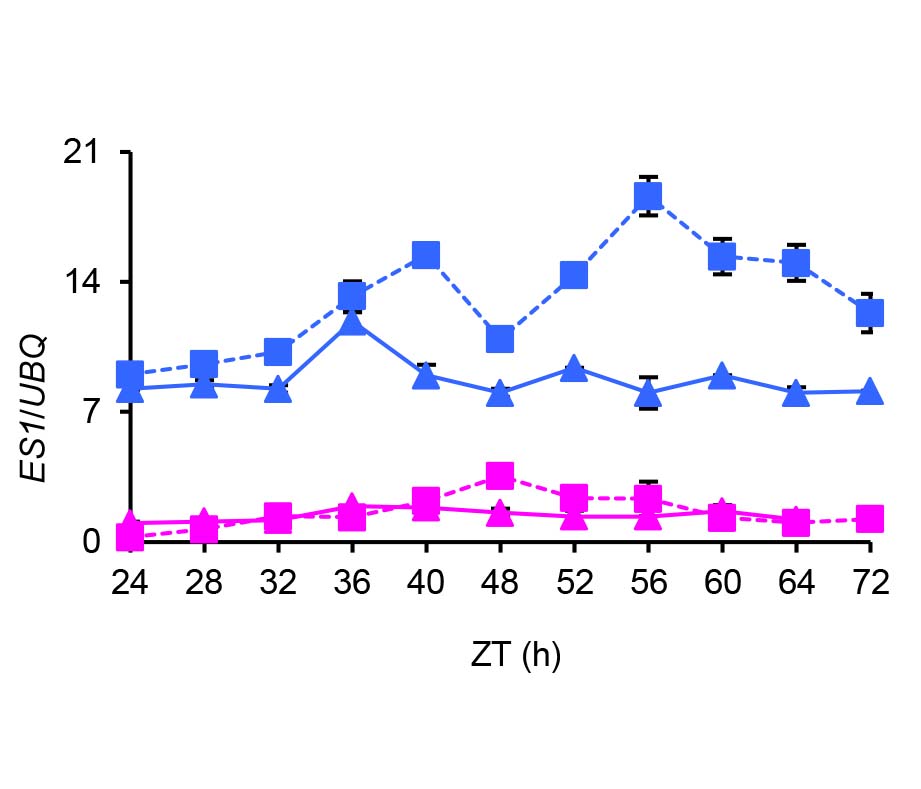
Jiangmin Xu , Dali Zeng , Miaomiao Huang , Qiaoli Fu , Hongzhen Jiang , Yuchun Rao , Han Lin . EARLY SENESCENCE 1 Participates in the Expression Regulation of Circadian Clock Genes and Response to Stress in Rice[J]. Chinese Bulletin of Botany, 2016 , 51(6) : 743 -756 . DOI: 10.11983/CBB16019
[1]Baba-Kasai A, Hara N, Takanom (2014).Tissue-specific and light-dependent regulation of phytochrome gene in rice. Plant Cell Environ 37, 2654–2666.
[2]Bae G, Choi G (2008).Decoding of light signals by plant phytochromes and their interac-
[3]ting proteins.Annu Rev Plant Biol 59, 281–311.
[4]Bai J, Zhu X, Wang Q, Zhang J, Chen H, Dong G, Zhu L, Zheng H, Xie Q, Nian J, Chen F, Fu Y, Qian Q, Zuo J (2015).Rice TUTOU1 encodes a suppressor of cAMP receptor-Like protein that is important for actin organization and panicle development. Plant Physiol 169, 1179–1191.
[5]Bendix C, Marshall CM, Harmon FG (2015).Circadian Clock Genes Universally Control Key Agricultural Traits. Mol Plant 8, 1135–1152.
[6]Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA (2008).Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol 147, 263–279.
[7]Bousset K, Oelgeschl?ger MH, Henriksson M, Schreek S, Burkhardt H, Litchfield DW, Lüscher-Firzlaff JM, Lüscher B (1994).Regulation of transcription factors c-Myc, Max, and c-Myb by casein kinase II. Cell Mol Biol Res 40, 501–511.
[8]Covington MF, Harmer SL (2007).The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5, e222.
[9]Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL (2008).Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9, R130.
[10]Daniel X, Sugano S, Tobin EM (2004).CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA 101, 3292–3297.
[11]Dong MA, Farré EM, Thomashow MF (2011).CIRCADIAN CLOCK-ASSOCIATED 1 and LATE ELONGATED HYPOCOTYL regulate of the C-REPEAT BINDING FACTOR (CBF) pathway in Arabidopsis. Proc Natl Acad Sci USA 108, 7241–7246.
[12]Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010).MYB transcription factors in Arabidopsis. Trends Plant Sci 15, 573–581.
[13]Farre EM, Liu T (2013).The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr opin plant bio 16, 621–629.
[14]Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D (1991).Light directed, spatially addressable parallel chemical synthesis. Science 251, 767–773.
[15]Fornara F, de Montaigu A, Sánchez-Villarreal A, Takahashi Y, Ver Loren van Themaat E, Huettel B, Davis SJ, Coupland G (2015).The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J 81, 695–706.
[16]Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999).Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582.
[17]Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012).Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109, 3167–3172.
[18]Green RM, Tobin EM (2002).The role of CCA1 and LHY in the plant circadian clock. Dev cell 2, 516–518.
[19]Green RM, Tingay S, Wang ZY, Tobin EM (2002).Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol 129, 576–584.
[20]Grundy J, Stoker C, Carré IA (2015).Circadian regulation of abiotic stress tolerance in plants. Front Plant Sci 27, 648.
[21]Harmer SL (2009).The circadian system in higher plants. Annu Rev Plant Biol 60, 357–377.
[22]Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012).Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79.
[23]Ito S, Nakamichi N, Nakamura Y, Niwa Y, Kato T, Murakami M, Kita M, Mizoguchi T, Niinuma K, Yamashino T, Mizuno T (2007).Genetic linkages between circadian clock-associated components and phytochrome-dependent red light signal transduction in Arabidopsis thaliana. Plant Cell Physiol 48, 971–983.
[24]Izawa T, Mihara M, Suzuki Y, Gupta M, Itoh H, Nagano AJ, Motoyama R, Sawada Y, Yano M, Hirai MY, Makino A, Nagamura Y (2011).Os-GIGANTEA confers robust diu-
[25]rnal rhythms on the global transcriptome of rice in the field.Plant Cell 23, 1741–1755.
[26]Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ (2009).Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21, 2624–2641.
[27]Kawamura M, Ito S, Nakamichi N, Yamashino T, Mizuno T (2008).The function of the clock-associated transcriptional regulator CCA1(CIRCADIAN CLOCK-ASSOCIATED 1) in Arabidopsis thaliana. Biosci Biotech Bioch 72, 1307–1316.
[28]Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JH, Dijkwel PP (2012).CADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci USA 109, 17129–17134.
[29]Liu T, Newton L, Liu MJ, Shiu SH, Farre EM (2016).A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol 170, 528-539.
[30]Liu Z, Liu Y, Pu Z, Wang J, Zheng Y, Li Y, Wei Y (2013).Regulation, evolution, and functionality of flavonoids in cereal crops. Biotechnol Lett 35, 1765–1780.
[31]Livak KJ, Schmittgen TD (2001).Analysis of relative gene data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408.
[32]Matsushika A, Makino S, Kojima M, Mizuno T (2000).Circadian waves of of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41, 1002–1012.
[33]McClung CR (2006).Plant circadian rhythms. Plant Cell 18, 792–803.
[34]Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song H-R, Carre′ IA, Coupland G (2002).LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2, 629–641.
[35]Mulekar JJ, Huq E (2014).Expanding roles of protein kinase CK2 in regulating plant
[36]growth and development.J Exp Bot 65, 2883–2893.
[37]Murakami M, Tago Y, Yamashino T, Mizuno T (2007).Comparative overviews of clock-
[38]associated genes of Arabidopsis thaliana and Oryza sativa.Plant cell physiol 48, 110–121.
[39]Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010).PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605.
[40]Niwa Y, Ito S, Nakamichi N, Mizoguchi T, Niinuma K, YamashinoT, MizunoT (2007).Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol 48, 925–937.
[41]Penfield S, Hall A (2009).A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 21, 1722–1732.
[42]Poethig RS (2003).Phase change and the regulation of developmental timing in plants. Science 301, 334–336.
[43]Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ (2012).The clock gene circuit in Arabidopsis includes a repressilator with additional feedback
[44]loops.Mol Syst Biol 8, 574.
[45]Pruneda-Paz JL, Breton G, Para A, Kay SA (2009).A functional genomics approach reveals CHE as a novel component of the Arabidopsis circadian clock. Science 323, 1481–1485.
[46]Rao Y, Yang Y, Xu J, Li X, Leng Y, Dai L, Huang L, Shao G, Ren D, Hu J, Guo L, Pan J, Zeng D (2015).EARLY SENESCENCE1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiol 169, 1225–1239.
[47]Riboni M, Galbiati M, Tonelli C, Conti L (2013).GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR
[48]OF OVEREXPRESSION OF CONSTANS1.Plant Physiol 162, 1706–1719.
[49]Sawa M, Kay SA(2011).GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci USA 108, 11698–11703.
[50]Schena M, Shalon D, Davis RW, Brown PO (1995).Quantitative monitoring of gene patterns with a complementary DNA microarray. Science 270, 467–470.
[51]Song YH, Ito S, Imaizumi T (2010).Similarities in the circadian clock and photoperiodism in plants. Curr Opin Plant Biol 13, 594–603.
[52]Syed NH, Prince SJ, Mutava RN, Patil G, Li S, Chen W, Babu V, Joshi T, Khan S, Nguyen HT (2015).Core clock, SUB1, and ABAR genes mediate flooding and drought responses via alternative splicing in soybean. J Exp Bot 66, 7129–7149.
[53]Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, Shinomura T (2005).Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17, 3311–3325.
[54]Takano M, Inagaki N, Xie X, Kiyota S, Baba-Kasai A, Tanabata T, Shinomura T (2009).Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice. Proc Natl Acad Sci USA 106, 14705–14710.
[55]Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004).Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006.
[56]Vilela B, Nájar E, Lumbreras V, Leung J, Pagès M (2015).Casein kinase 2 negatively regulates abscisic acid-activated SnRK2s in the core abscisic acid-signaling module. Mol Plant 8, 709–721.
[57]王颖姮, 邓其明, 李平 (2006).基因芯片技术在水稻研究中的应用. 中国农学通报 8, 55–59.
[58]Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X (2012).Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44, 950–954.
[59]Woychik RP, Klebig ML, Justice MJ, Magnuson TR, Avner ED (1998).Functional genomics in the post-genome era. Mutat Res 400, 3–14.
[60]Zhang L, Liu G, Zhao G, Xia C, Jia J, Liu X, Kong X (2014).Characterization of a wheat R2R3-MYB transcription factor gene, TaMYB19, involved in enhanced abiotic stresses in Arabidopsis. Plant Cell Physiol 10, 1802–1812.
/
| 〈 |
|
〉 |