1 光敏色素调控植物发育
2 光敏色素磷酸化修饰在光信号通路中的作用
2.1 phyA的磷酸化与去磷酸化作用
Figure 1 Multiple sequence alignments of monocot and dicot phyA proteinsAt: Arabidopsis thaliana phyA; As: Avena sativa phyA; As: Avena sativa phyA type 3; Os: Oryza sativa phyA; Hv: Hordeum vulgare subsp. vulgare phyA; Ta: Triticum aestivum phyA type 1; Bd: Brachypodium distachyon phyA type 3-like; Zm: Zea mays phyA; Sb: Sorghum bicolor phyA; Sp: Sorghum propinquum phyA; St: Solanum tuberosum phyA. * indicates phosphorylation sites at Ser8, Ser18 and Ser598 identified in oat phyA 图1 单子叶和双子叶植物中phyA蛋白的多序列对比 At: 拟南芥phyA; As: 燕麦phyA; As3: 燕麦phyA 3型; Os: 水稻phyA; Hv: 大麦亚种phyA; Ta: 小麦phyA 1型; Bd: 二穗短柄草phyA 3型; Zm: 玉米phyA; Sb: 高粱phyA; Sp: 拟高粱phyA; St: 马铃薯phyA。*表示燕麦phyA中已鉴定的磷酸化位点Ser8、Ser18和Ser598 |
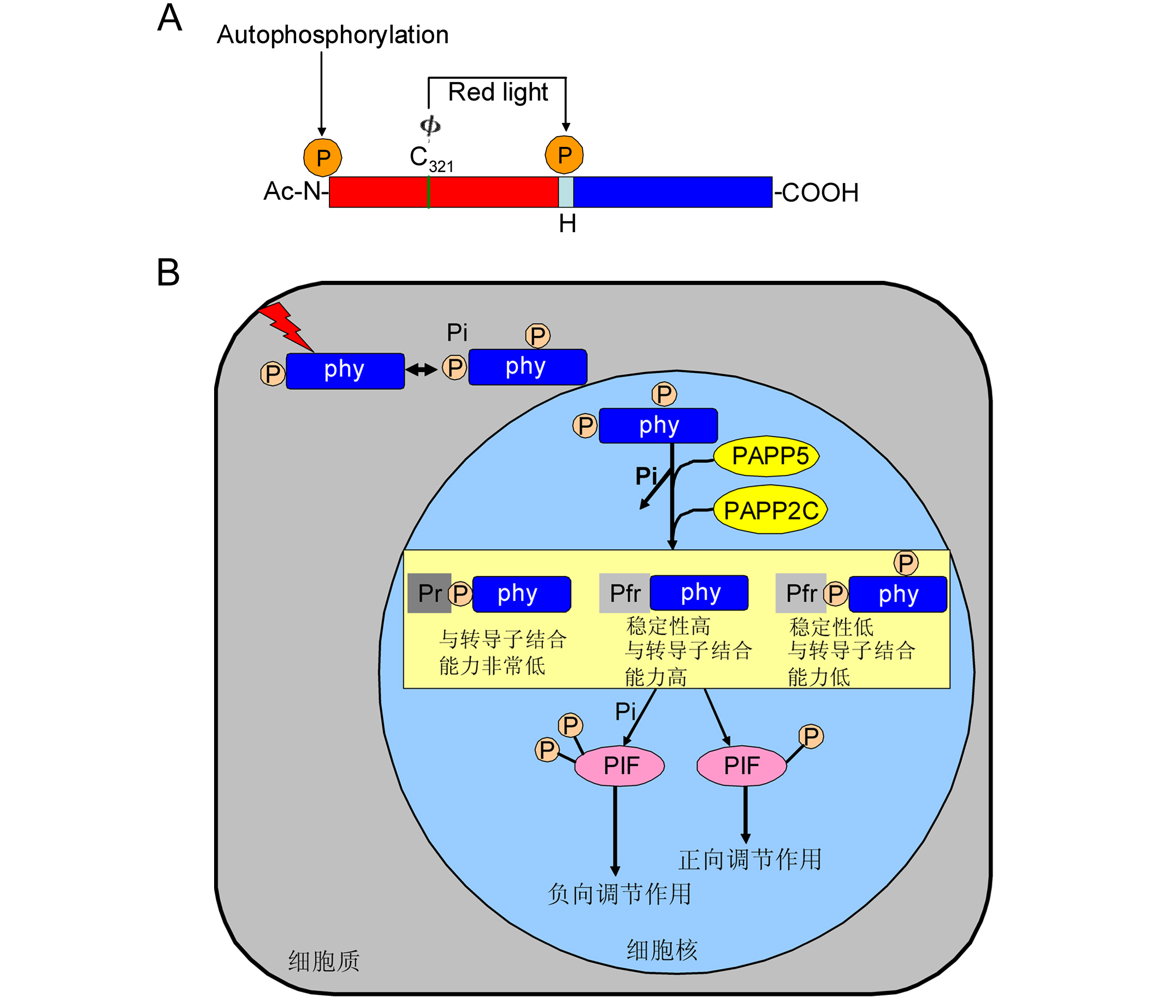
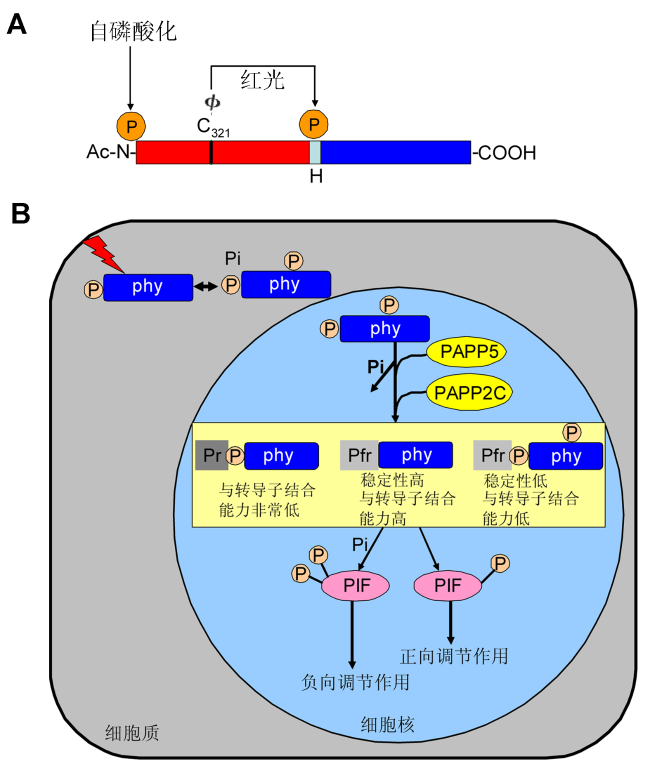
Figure 2 A proposed model of phosphorylation modification in the phytochrome signaling pathway (Ryu et al., 2005; Galvão et al., 2012)(A) Domain structure and phosphorylation sites in oat phyA. The N-terminal (N) and C-terminal domains (COOH) are shown by rectangles. Φ indicates phytochromobilin, cova- lently attached to Cys321 (C321). The middle of the small rectangle represents “hinge region (H)”, the phosphorylation site at Ser598 is shown by arrow in the middle. Three types of posttranslational modification of oat phyA have been reported previously: N-acetylation (ac-N), phytochromobilin ligation to Cys321 and phosphorylation in the N-terminal region at Ser7. Phosphorylation at the N-terminus was suggested to be the site of phytochrome “autophosphorylation” (Lapko et al., 1999); (B) Light triggers photoconversion of the Pr-phyto- chromes to the Pfr-phytochromes, which initiates the phyto- chrome-mediated photosignaling. Pfr-phytochromes are pho- sphorylated by their intrinsic kinase activity as well as by phytochromes-associated kinase(s) and are reversibly dephosphorylated by phosphatase, such as, PAPP5, PAP- P2C. The Pfr-phytochromes dephosphorylates in the N-ter- minal extension and the hinge region is relieved from phosphory- lation mediated destabilization, exhibiting a high affinity to signal transducers. However, the unphosphorylated Pfr-phytochromes were not. Furthermore, the phosphorylated Pr-phytochromes possess a very lower affinity toward signal transducers. These mechanisms lead to the existence of two state of PIF. One is PIF was phosphorylated by phytochromes results in the negative regulation of light signaling. Another is phytochromes were dephosphorylated and could not phosphorylate PIF, which results in the positive regulation of light signaling. 图2 光敏色素信号通路的磷酸化修饰模式图(Ryu et al., 2005; Galvão et al., 2012) (A) 燕麦phyA结构域和磷酸化位点。N端结构域(N)和C端结构域(COOH)用矩形表示, Φ表示植物后胆色素, 它与氨基酸Cys321 (C321)共价连接。中间的小矩形表示铰链区(H), 中间箭头指示的位置为铰链区域一个磷酸化位点Ser598。目前已经报道燕麦phyA有3种翻译后修饰: N-乙酰化(Ac-N), 植物后胆色素连接到phyA 321位的半胱氨酸上以及磷酸化N端区域的第7位丝氨酸。光敏色素的自磷酸化位点在N端区域(Lapko et al., 1999); (B) 光促使无活性的光敏色素转变为有活性的光敏色素, 从而开启了光敏色素调节的光信号, 有活性的光敏色素可以自磷酸化以及被一些光敏色素相关的激酶磷酸化。磷酸化的光敏色素会被一些磷酸酶(PAPP5和PAPP2C)去磷酸化。有活性的光敏色素在N端和铰链区被去磷酸化从而缓解了磷酸化介导的去稳定作用, 进而与下游信号分子结合能力增强, 而没有磷酸化的有活性的光敏色素反之。此外, 磷酸化的没有活性的光敏色素与下游信号分子结合能力非常低。这些机制导致下游PIF存在2种状态, 一种是PIF被光敏色素磷酸化从而负向调控光信号; 另一种是光敏色素由于被去磷酸化所以不能磷酸化PIF, 最终导致正向调节光信号。 |
2.2 phyB的磷酸化与去磷酸化作用
Figure 3 Multiple sequence alignments of monocot and dicot phyB proteinsAt: Arabidopsis thaliana phyB; As: Avena sativa phyB; Os: Oryza sativa phyB; Hv: Hordeum vulgare subsp. vulgare phyB; Ta: Triticum aestivum putative phyB; Bd: Brachypodium distachyon phyB-like; Zm: Zea mays phyB 2; Sb: Sorghum bicolor phyB; Sp: Sorghum propinquum phyB; St: Solanum tuberosum phyB. * indicates phosphorylation sites at Ser86 and Tyr104 identified in Arabidopsis phyB 图3 单子叶和双子叶植物中phyB蛋白的多序列对比 At: 拟南芥phyB; As: 燕麦phyB; Os: 水稻phyB; Hv: 大麦亚种phyB; Ta: 小麦phyB; Bd: 二穗短柄草类phyB; Zm: 玉米phyB 2; Sb: 高粱phyB; Sp: 拟高粱phyB; St: 马铃薯phyB。*表示拟南芥phyB中已鉴定的磷酸化位点Ser86和Tyr104 |


 Home
Home