

Current Research Advances on Glutamate Receptors (GLRs) in Plants
# Co-first authors
Received date: 2015-12-03
Accepted date: 2016-04-01
Online published: 2016-12-02
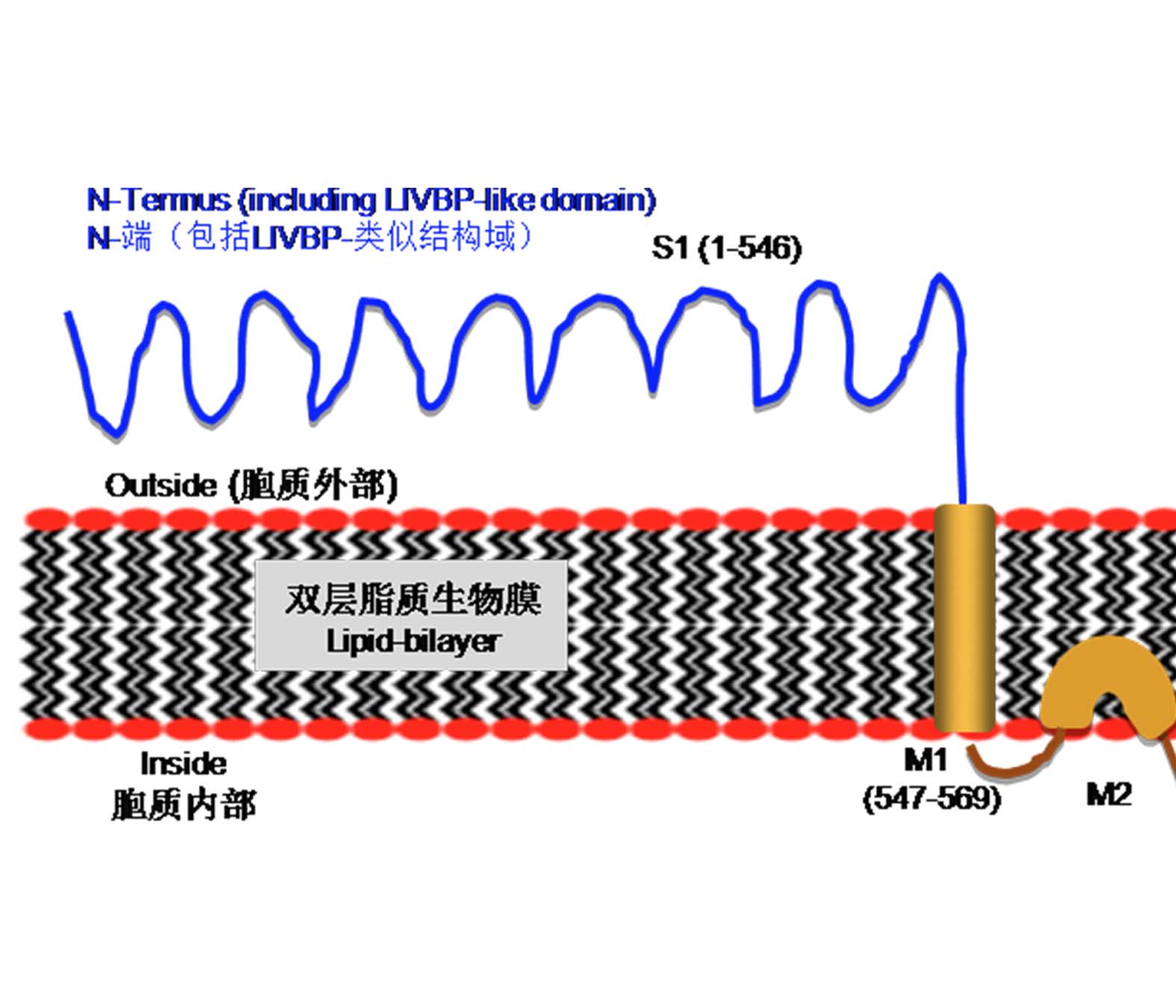
In mammals, ionotropic glutamate receptors (iGLuR) are amino acids (e.g. glutamate and glycine) -gated cation channels, and exhibit molecular functions in the regulation of excitatory neurotransmission as well as in directing neuron growth. Since 1998 twenty genes homologous to iGLuR have been identified in Arabidopsis genome (termed AtGLRs), with reported functions involved in many biological processes including light signaling, root-tip meristematic cell activity, pollen tube growth, cytosolic calcium ion flux and response to varied biotic and abiotic stresses. This paper comprehensively summarizes research achievements or advances in terms of plant glutamate receptors and amino acid (e.g. glutamate) signaling in the past more than ten years, with major issues focusing on e.g. the protein structure of GLRs, a relationship between activation of ion channels and their ligands, their gene expression patterns as well as possible biological roles in plants, thus hopefully providing valuable information for researchers related to this field.
Yichen Sun , Yongkun Feng , Laihua Liu , Mingjie He , Yiyin Chen , Fengqiu Cao , Diqin Li , Chao Yang , Xiaoyuan Cheng , Lu Liu , Dongxue Shi , Tengfei Fan . Current Research Advances on Glutamate Receptors (GLRs) in Plants[J]. Chinese Bulletin of Botany, 2016 , 51(6) : 827 -840 . DOI: 10.11983/CBB15212
| [1] | Aouini A, Matsukura C, Ezura H, Asamizu E (2012). Characterization of 13 glutamate receptor-like genes encoded in the tomato genome by structure phylogeny and expression profiles.Gene 493, 36-43. |
| [2] | Ayalon G, Segev E, Elgavish S, Stern-Bach Y (2005). Two regions in the N-terminal domain of ionotropic glutamate receptor 3 form the subunit oligomerization interfaces that control subtype-specific receptor assembly.J Biol Chem 280, 15053-15060. |
| [3] | Ayalon G, Stern-Bach Y (2001). Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions.Neuron 31, 103-113. |
| [4] | Brenner ED, Martinez-Barboza N, Clark AP, Liang QS, Stevenson DW, Coruzzi GM (2000). Arabidopsis mutants resistant to S(+)-β-methyl-α, β-diaminopropionic acid, a cycad-derived glutamate receptor agonist.Plant Physiol 124, 1615-1624. |
| [5] | Chávez AE, Singer JH, Diamond JS (2006). Fast neuro- transmitter release triggered by Ca2+ influx through AMPA- type glutamate receptors.Nature 443, 705-708. |
| [6] | Chen GQ, Cui CH, Mayer ML, Gouaux E (1999). Functional characterization of a potassium-selective prokaryotic glu- tamate receptor.Nature 402, 817-821. |
| [7] | Chiu JC, Brenner ED, Desalle R, Nitabach MN, Holmes TC, Coruzzi GM (2002). Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana.Mol Biol Evol 19, 1066-1082. |
| [8] | Chiu JC, Desalle R, Lam HM, Meisel L, Coruzzi G (1999). Molecular evolution of glutamate receptors: a primitive signaling mechanism that existed before plants and animals diverged.Mol Biol Evol 16, 826-838. |
| [9] | Cho D, Kim SA, Murata Y, Lee S, Jae SK, Nam HG, Kwak JM (2009). De-regulated expression of the plant gluta- mate receptor homolog AtGLR3.1 impairs long-term Ca2+ programmed stomatal closure.Plant J 58, 437-449. |
| [10] | Davenport R (2002). Glutamate receptors in plants.Ann Bot 90, 549-557. |
| [11] | Dennison KL, Spalding EP (2000). Glutamate-gated calci- um fluxes in Arabidopsis.Plant Physiol 124, 1511-1514. |
| [12] | Dingledine R, Borges K, Bowie D, Traynelis SF (1999). The glutamate receptor ion channels. Pharmacol Rev 51, 7-61. |
| [13] | Dubos C, Huggins D, Grant GH, Knight MR, Campbell MM (2003). A role for glycine in the gating of plant NMDA- like receptors.Plant J 35, 800-810. |
| [14] | Dubos C, Willment J, Huggins D, Grant GH, Campbell MM (2005). Kanamycin reveals the role played by glutamate receptors in shaping plant resource allocation.Plant J 43, 348-355. |
| [15] | Forde BG, Cutler SR, Zaman N, Krysan PJ (2013). Gluta- mate signaling via a MEKK1 kinase-dependent pathway induces changes in Arabidopsis root architecture.Plant J 75, 1-10. |
| [16] | Kang J, Sohum M, Turano FJ (2004). The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss.Plant Cell Physiol 45, 1380-1389. |
| [17] | Kang J, Turano FJ (2003). The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana.Proc Natl Acad Sci USA 100, 6872-6877. |
| [18] | Kang S, Kim HB, Lee H, Choi JY, Heu S, Oh CJ, Kwon SI, An CS (2006). Overexpression in Arabidopsis of a plasma membrane-targeting glutamate receptor from small radish increases glutamate-mediated Ca2+ influx and delays fun- gal infection.Mol Cell 21, 418-427. |
| [19] | Kim SA, Kwak JM, Jae SK, Wang MH, Nam HG (2001). Overexpression of the AtGluR2 gene encoding an Arabi- dopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants.Plant Cell Physiol 42, 74-84. |
| [20] | Kong DD, Ju CL, Parihar A, Kim S, Cho D, Kwak JM (2015). Arabidopsis glutamate receptor homolog atglr3.5 modulates cytosolic Ca2+ level to counteract effect of abscisic acid in seed germination.Plant Physiol 167, 1630-1642. |
| [21] | Kushwaha R, Singh A, Chattopadhyay S (2008). Calmo- dulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development.Plant Cell 20, 1747-1759. |
| [22] | Lacombe B (2001). The identity of plant glutamate recap- tors.Science 292, 1486-1487. |
| [23] | Lam H, Chiu J, Hsieh M, Lee M, Oliveira IC, Shin M, Coruzzi G (1998). Glutamate-receptor genes in plants.Nature 396, 125-126. |
| [24] | Li HJ, Yang WC (2012). Emerging role of ER quality control in plant cell signal perception.Protein Cell 3, 10-16. |
| [25] | Li J, Zhu SH, Song XW, Shen Y, Chen HM, Yu J, Yi K, Liu YF, Karplus VJ, Wu P, Deng XW (2006). A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem.Plant Cell 18, 340-349. |
| [26] | Manzoor H, Kelloniemi J, Chiltz A, Wendehenne D, Pugin A, Poinssot B, Garcia-Brugger A (2013). Involvement of the glutamate receptor AtGLR3.3 in plant defense sig- naling and resistance to Hyaloperonospora arabidopsidis.Plant J 76, 466-480. |
| [27] | McFeeters RL, Oswald RE (2004). Emerging structural explanations of ionotropic glutamate receptor function.FASEB J 18, 428-438. |
| [28] | Meyerhoff O, Müller K, Roelfsema MR, Latz A, Lacombe B, Hedrich R, Dietrich P, Becker D (2005). AtGLR3.4, a glutamate receptor channel-like gene is sensitive to touch and cold.Planta 222, 418-427. |
| [29] | Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham MG, Liu LH, Obermeyer G, Feijó J (2011). Glutamate receptor-like genes form Ca2+ chan- nels in pollen tubes and are regulated by pistil D-serine.Science 332, 434-437. |
| [30] | Mousavi SAR, Chauvin A, Pascaud F, Kellenberger S, Farmer EE (2013). GLUTAMATE RECEPTOR-LIKE ge- nes mediate leaf-to-leaf wound signaling.Nature 500, 422-441. |
| [31] | Nagata T, Iizumi S, Satoh K, Ooka H, Kawai J, Carninci P, Hayashizaki Y, Otomo Y, Murakami K, Matsubara K, Kikuchi S (2004). Comparative analysis of plant and animal calcium signal transduction element using plant full-length cDNA data.Mol Biol Evol 21, 1855-1870. |
| [32] | Parsons CG, Panchenko VA, Pinchenko VO, Tsyndreko AY, Krishtal OA (1966). Comparative patch-clamp stud- ies with freshly dissociated rat hippocampal and striatal neurons on the NMDA receptor antagonistic effects of amantadine and memantin.Eur J Neurosci 8, 446-454. |
| [33] | Penn AC, Williams SR, Greger IH (2008). Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum.EMBO J 27, 3056-3068. |
| [34] | Price MB, Jelesko J, Okumoto S (2013). Glutamate receptor homologs in plants: functions and evolutionary origins.Front Plant Sci 3, 1-10. |
| [35] | Price MB, Kong DD, Okumoto S (2013). Inter-subunit in- teractions between glutamate-like receptors in Arabi- dopsis.Plant Signal Behav 8, e27034. |
| [36] | Qi Z, Stephens NR, Spalding EP (2006). Calcium entry me- diated by glr3.3, an Arabidopsis glutamate receptor with a broad agonist profile.Plant Physiol 142, 963-971. |
| [37] | Roy SJ, Gilliham M, Berger B, Essah PA, Cheffings C, Miller AJ, Davenport RJ, Liu LH, Skynner MJ, Davies JM, Richardson P, Leigh RA, Tester M (2008). Investi- gating glutamate receptor-like gene co-expression in Ara- bidopsis thaliana.Plant Cell Environ 31, 861-871. |
| [38] | Singh A, Kanwar P, Yadav AK, Mishra M, Jha SK, Baranwal V, Pandey A, Kappor S, Tyagi AK, Pandey GK (2014). Genome-wide expressional and functional analysis of calcium transport elements during abiotic stress and development in rice.FEBS J 281, 894-915. |
| [39] | Sivaguru M, Pike S, Gassmann W, Baskin TI (2003). Aluminum rapidly depolymerized cortical microtubules and depolarizes the plasma membrane: evidence that these responses are mediated by a glutamate receptor.Plant Cell Physiol 44, 667-675. |
| [40] | Sobolevsky AI, Rosconi MP, Gouaux E (2009). X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor.Nature 462, 745-756. |
| [41] | Stephens NR, Qi Z, Spalding EP (2008). Glutamate re- ceptor subtypes evidenced by differences in desensitiza- tion and dependence on the GLR3.3 and GLR3.4 genes.Plant Physiol 146, 529-538. |
| [42] | Tapken D, Anschütz U, Liu LH, Huelsken T, Seebohm G, Becker D, Hollmann M (2013). A plant homolog of animal glutamate receptors is an ion channel gated by multiple hydrophobic amino acids.Sci Signal 6, a47. |
| [43] | Tapken D, Hollmann M (2008). Arabidopsis thaliana gluta- mate receptor ion channel function demonstrated by ion pore transplantation.J Mol Biol 383, 36-48. |
| [44] | Teardo E, Carraretto L, Bortoli SD, Costa A, Behera S, Wagner R, Schiavo FL, Formentin E, Szabo I (2015). Alternative splicing-mediated targeting of the Arabidopsis GLUTAMATE RECEPTOR3.5 to mitochondria affects or- ganelle morphology1.Plant Physiol 167, 216-227. |
| [45] | Teardo E, Formentin E, Segalla A, Giacometti GM, Marin O, Zanetti MA, Schiavo FL, Zoratti M, Szabò I (2011). Dual localization of plant glutamate receptor AtGLR3.4 to plastids and plasmamembrane.Biochim Biophys Acta 1807, 359-367. |
| [46] | Teardo E, Segalla A, Formentin E, Zanetti M, Marin O, Giacometti GM, Schiavo FL, Zoratti M, Szabò I (2010). Characterization of a plant glutamate receptor activity.Cell Physiol Biochem 26, 253-262. |
| [47] | Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan HJ, Myers SJ, Dingledine R (2010). Glutamate receptor ion chan- nels: structure, regulation, and function.Pharmacol Rev 62, 405-496. |
| [48] | Turano FJ, Muhitch MJ, Felker FC, McMahon MB (2002). The putative glutamate receptor 3.2 from Arabidopsis thaliana (AtGLR3.2) is an integral membrane peptide that accumulates in rapidly growing tissues and persists in vascular-associated tissues.Plant Sci 163, 43-51. |
| [49] | Turano FJ, Panta GR, Allard MW, Berkum PV (2001). The putative glutamate receptors from plants are related to two superfamilies of animal neurotransmitter receptors via distinct evolutionary mechanisms.Mol Biol Evol 18, 1417-1420. |
| [50] | Ulbrich MH, Isacoff EY (2008). Rules of engagement for NMDA receptor subunits.Proc Natl Acad Sci USA 105, 14163-14168. |
| [51] | Vatsa P, Chiltz A, Bourque S, Wendehenne D, Garcia- Brugger A, Pugin A (2011). Involvement of putative glutamate receptors in plant defence signaling and NO production.Biochimie 93, 2095-2101. |
| [52] | Vincill ED, Bieck AM, Spalding EP (2012). Ca2+ conduction by an amino acid-gated ion channel related to glutamate receptors.Plant Physiol 159, 40-46. |
| [53] | Vincill ED, Clarin AE, Molenda JN, Spalding EP (2013). Interacting glutamate receptor-like proteins in phloem re- gulate lateral root initiation in Arabidopsis.Plant Cell 25, 1304-1313. |
| [54] | Walch-Liu P, Forde BG (2008). Nitrate signaling mediated by the NRT1.1 nitrate transporter antagonises L-gluta- mate-induced changes in root architecture.Plant J 54, 820-828. |
| [55] | Walch-Liu P,Liu LH, Remans T, Tester M, Forde BG (2006). Evidence that L-glutamate can act as an exoge- nous signal to modulate root growth and branching in Arabidopsis thaliana.Plant Cell Physiol 47, 1045-1057. |
/
| 〈 |
|
〉 |