

Mechanism of The “Pioneer” Transcription Factor LEC1 in Resetting Vernalized State in Early Embryos
Received date: 2017-12-04
Accepted date: 2018-01-10
Online published: 2018-01-10
Flowering is an important process for plants to switch from vegetative to reproductive phase. Vernalization is a process whereby plants acquire the ability to flower after exposure to a prolonged cold temperature. In Arabidopsis, inhibitor-type transcription factor FLOWERING LOCUS C (FLC) is a critical point in vernalization-mediated flowering pathway. Previous studies in Arabidopsis thaliana revealed that two homologous epigenome readers, VAL1 and VAL2, re- cognize a cis DNA element in the nucleation region for Polycomb group (PcG) silencing at the key floral repressor FLC, engaging Polycomb group proteins to induce epigenetic silencing of FLC by histone 3 lysine trimethylation (H3K27me3) during vernalization. This silencing is maintained in subsequent growth and development under normal temperature, namely vernalization memory. How to delete vernalization memory in the next generation to de novo activate FLC expression, preventing the offspring from flowering before or during winter, is not clear. Recently, Chinese scientist have found that a seed-specific transcription factor LEAFY COTYLEDON1 (LEC1) functions in deleting vernalization memory and reactivating the expression of FLC in the pro-embryo by resetting the chromatin states from the silenced state (marked by H3K27me3) to an active state (H3K36me3). This study provides important understanding of molecular and genetic mechanisms for flowering control by vernalization, and a novel strategy to genetically manipulate crop flowering times for the benefit of agricultural production, which is a great breakthrough of this field.
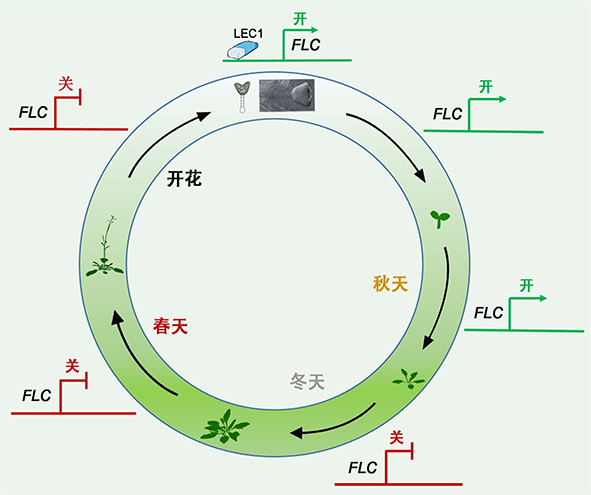
Xu Shujuan , Chong Kang . Mechanism of The “Pioneer” Transcription Factor LEC1 in Resetting Vernalized State in Early Embryos[J]. Chinese Bulletin of Botany, 2018 , 53(1) : 1 -4 . DOI: 10.11983/CBB17234
| [1] | Crevillén P, Yang HC, Cui X, Greeff C, Trick M, Qiu Q, Cao XF, Dean C (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state.Nature 515, 587-590. |
| [2] | Csorba T, Questa JI, Sun Q, Dean C (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA 111, 16160-16165. |
| [3] | Dubcovsky J, Loukoianov A, Fu DL, Valarik M, Sanchez A, Yan LL (2006). Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol Biol 60, 469-480. |
| [4] | Fornara F, de Montaigu A, Coupland G (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550-550. e2 |
| [5] | He YH (2012). Chromatin regulation of flowering.Trends Plant Sci 17, 556-562. |
| [6] | Heo JB, Sung S (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA.Science 331, 76-79. |
| [7] | Hepworth J, Dean C (2015). Flowering Locus C's lessons: conserved chromatin switches underpinning developmental timing and adaptation.Plant Physiol 168, 1237-1245. |
| [8] | Kim DH, Doyle MR, Sung S, Amasino RM (2009). Vernalization: winter and the timing of flowering in plants.An- nu Rev Cell Dev Biol 25, 277-299. |
| [9] | Kippes N, Debernardi JM, Vasquez-Gross HA, Akpinar BA, Budak H, Kato K, Chao S, Akhunov E, Dubcovsky J (2015). Identification of the VERNALIZATION 4 gene reveals the origin of spring growth habit in ancient wheats from South Asia. Proc Natl Acad Sci USA 112, E5401-E5410. |
| [10] | Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for em- bryo development.Plant Cell 15, 5-18. |
| [11] | Qüesta JI, Song J, Geraldo N, An HL, Dean C (2016). Arabidopsis transcriptional repressor VAL1 triggers Poly- comb silencing at FLC during vernalization. Science 353, 485-488. |
| [12] | Tao Z, Shen LS, Gu XF, Wang YZ, Yu H, He YH (2017). Embryonic epigenetic reprogramming by a pioneer trans- cription factor in plants.Nature 551, 124-128. |
| [13] | Xiao J, Xu SJ, Li CH, Xu YY, Xing LJ, Niu YD, Huan Q, Tang YM, Zhao CP, Wagner D, Gao CX, Chong K (2014). O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during ver- nalization in winter wheat. Nat Commun 5, 4572. |
| [14] | Xing LJ, Li J, Xu YY, Xu ZH, Chong K (2009). Phosphorylation modification of wheat lectin VER2 is associated with vernalization-induced O-GlcNAc signaling and intracellular motility. PLoS One 4, e4854. |
| [15] | Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103, 19581-19586. |
| [16] | Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003). Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100, 6263-6268. |
| [17] | Yong WD, Xu YY, Xu WZ, Wang X, Li N, Wu JS, Liang TB, Chong K, Xu ZH, Tan KH, Zhu ZQ (2003). Vernalization-induced flowering in wheat is mediated by a lectin-like gene VER2. Planta 217, 261-270. |
| [18] | Yuan WY, Luo X, Li ZC, Yang WN, Wang YZ, Liu R, Dui JM, He YH (2016). A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat Genet 48, 1527-1534. |
/
| 〈 |
|
〉 |