

Identification of the DEAD-box Gene Family and Expression Pattern of the StRH10 Gene Under Salt Stress in Scaevola taccada
1College of Life and Environmental Sciences, Wenzhou University, Wenzhou 325035, China; 2College of Life Science and Technology, Lingnan Normal University, Zhanjiang Key Laboratory of Mangrove Ecosystem Conservation and Restoration, Zhanjiang 524048, China
Received date: 2025-06-24
Revised date: 2025-08-04
Online published: 2025-09-05
INTRODUCTION: The global area of saline-alkali land is approximately 932 million hectares. The problem of soil salinization caused by seawater intrusion in china's coastal areas is intensifying. Salt stress is an important abiotic stress that affects plant growth and development. Throughout the long-term evolutionary process, plants have developed complex adaptation mechanisms to cope with stress, with gene expression regulation playing a central role. Among the numerous salt stress-responsive genes, the DEAD-box RNA helicase gene family has garnered significant attention due to its crucial roles in plant growth, development, and abiotic stress responses. Studies have confirmed that members of this family are involved in the regulation of salt stress in a variety of plants. Therefore, the DEAD-box RNA helicase gene family is an important object for studying the molecular mechanism of plant salt stress response.
RATIONALE: Scaevola taccada is a semi-mangrove plant widely distributed in tropical and subtropical coastal areas. It exhibits excellent salt tolerance, drought resistance, and resilience to wind and waves. This plant holds significant ecological value for coastal sand stabilization and ecological restoration. Although the DEAD-box RNA helicase gene has been extensively studied in model plants, the genome-wide identification, chromosomal distribution, and salt stress response mechanism of this gene family remain unclear in S. taccada. Therefore, based on the genome data of S. taccada, the members of the DEAD-box gene family (StRHs) were systematically identified, and the tissue expression specificity of the StRH10 gene and its response pattern under salt stress were analyzed, aiming to provide a preliminary scientific basis for the molecular breeding of high-salt tolerant S. taccada seedlings.
RESULTS: Fifty StRH genes were identified from the genome of S. taccada. Most StRH proteins contain 10 conserved motifs, while a few contain 8 or 9. The StRH proteins comprise 18 conserved domains, primarily SrmB and DEAD. Seven StRH genes lack introns. The promoter region of the StRH genes is rich in cis-acting elements associated with the stress response. S. taccada and Arabidopsis thaliana have 20 pairs of collinear genes, and S. taccada and Helianthus annuus have 39 pairs of collinear genes. StRH10 protein is most closely related to the chloroplast-localized DEAD-box RNA helicase AtRH03 protein in A. thaliana. The subcellular localization prediction indicated that the StRH10 protein is localized to the chloroplasts. StRH10 gene was the most expressed in leaves. Under salt stress treatment, the expression level of the StRH10 gene did not show a significant difference with increasing NaCl concentration but was significantly up-regulated when the NaCl concentration reached 400 mmol·L-1.
CONCLUSION: The 50 members of the DEAD-box gene family identified in the genome of S. taccada in this study were unevenly distributed across the chromosomes. Most StRH proteins contain 10 conserved motifs and are clustered within the same evolutionary branch. The promoter region of StRH genes is rich in stress-responsive cis-acting elements such as ABRE, TC-rich repeats, MBS, LTR, WUN-motif and ARE. Collinearity analysis showed that there were 39 pairs of collinearity genes in S. taccada and H. annuus. The StRH10 gene, which encodes a protein predicted to be located in the chloroplast, exhibits high expression in leaves.Under salt stress, the expression of the StRH10 gene was significantly up-regulated only when the NaCl concentration reached 400 mmol·L-1, indicating that this gene responds to salt stress with a certain concentration threshold.
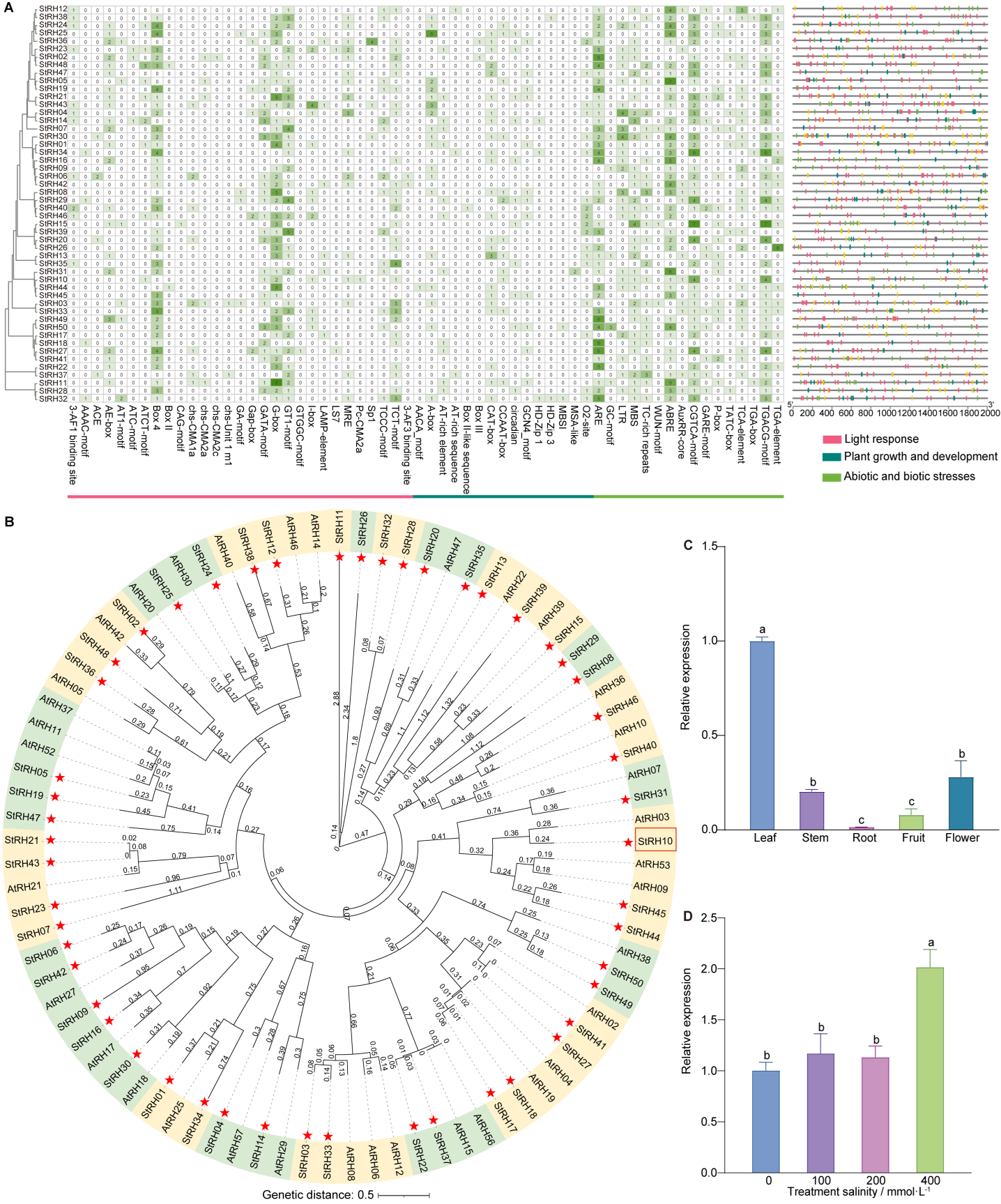
(A) DEAD-box gene family Cis-acting elements in the promoter region; (B) Phylogenetic tree of DEAD-box gene family members in S. taccada and A. thaliana (The red pentagram indicates the RH gene of S. taccada); (C) StRH10 expression levels in different organizations; (D) StRH10 expression levels under salt stress
Key words: Scaevola taccada; DEAD-box gene family; StRH10 gene; Salt stress; Expression pattern
LIAO Li-Xian
,
LUO Jing-Lin
,
XIAN Yu-Ping
,
SANG Hai-Shou
,
ZHENG Chun-Fang
,
ZHANG Ying
.
Identification of the DEAD-box Gene Family and Expression Pattern of the StRH10 Gene Under Salt Stress in Scaevola taccada[J]. Chinese Bulletin of Botany, 0
/
| 〈 |
|
〉 |