

Received date: 2025-05-26
Revised date: 2025-07-25
Online published: 2025-09-03
INTRODUCTION: As an important ornamental species, chrysanthemum (Chrysanthemum morifolium Ramat.) depends on precise flowering time regulation, which is a key factor affecting its economic value and market demand. The bud sport of cut chrysanthemum cultivar 'Jinshan' maintains vegetative growth under normal cultivation conditions and fails to initiate floral transition. However, the underlying regulatory mechanisms responsible for its flowering defect remain unclear.
RATIONALE: To explore the molecular basis of the flowering inhibition in the ‘Jinshan’ bud sport, we conducted a comparative analysis using both the wild-type and the bud sport lines. Sequence-related amplified polymorphism (SRAP) molecular markers were used to evaluate their genetic backgrounds. In addition, transcriptome sequencing (RNA-seq) was performed on leaf and shoot apical meristem (SAM) tissues to identify differentially expressed genes (DEGs) associated with floral transition.
RESULTS: SRAP analysis revealed a high degree of genomic similarity between the bud sport and wild-type lines, suggesting that large-scale genetic structural variations are unlikely to be the cause of the floral defect. Transcriptomic analysis identified a substantial number of DEGs between the two lines. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses indicated significant enrichment of plant hormone signal transduction pathways in both leaf and SAM tissues. Further analysis of flowering-related regulatory networks revealed widespread transcriptional dysregulation. In the photoperiod pathway, abnormal expression of the CO-FT module may have impaired the perception and transmission of flowering signals. In the autonomous pathway, upregulation of MAF2 (a homolog of FLC) and downregulation of FLD likely led to the repression of downstream floral integrators. In the gibberellin (GA) signaling pathway, a significant downregulation of the gibberellin receptor GID1, as well as the downregulation of the GA biosynthesis enzyme GA3ox and the upregulation of the GA deactivating enzyme GA2ox, may result in a reduction in active gibberellin content. In the early regulation network of flower organ formation, the expression of SOC1, AP1, FUL, AFL, and SPLs was significantly downregulated, and LFY expression in the apical meristem was nearly absent. These changes may collectively block floral primordium formation and flower organ differentiation in the bud sport.
CONCLUSION: This study reveals that the abnormal expression of multiple key genes in the photoperiod, autonomous, and gibberellin signaling pathways contribute to the failure of floral transition in the bud sport. These findings provide preliminary insights into the potential mechanism underlying flowering inhibition in the 'Jinshan' bud sport at the transcriptomic level, and provide theoretical support and candidate genes for molecular breeding of flowering time in chrysanthemum.
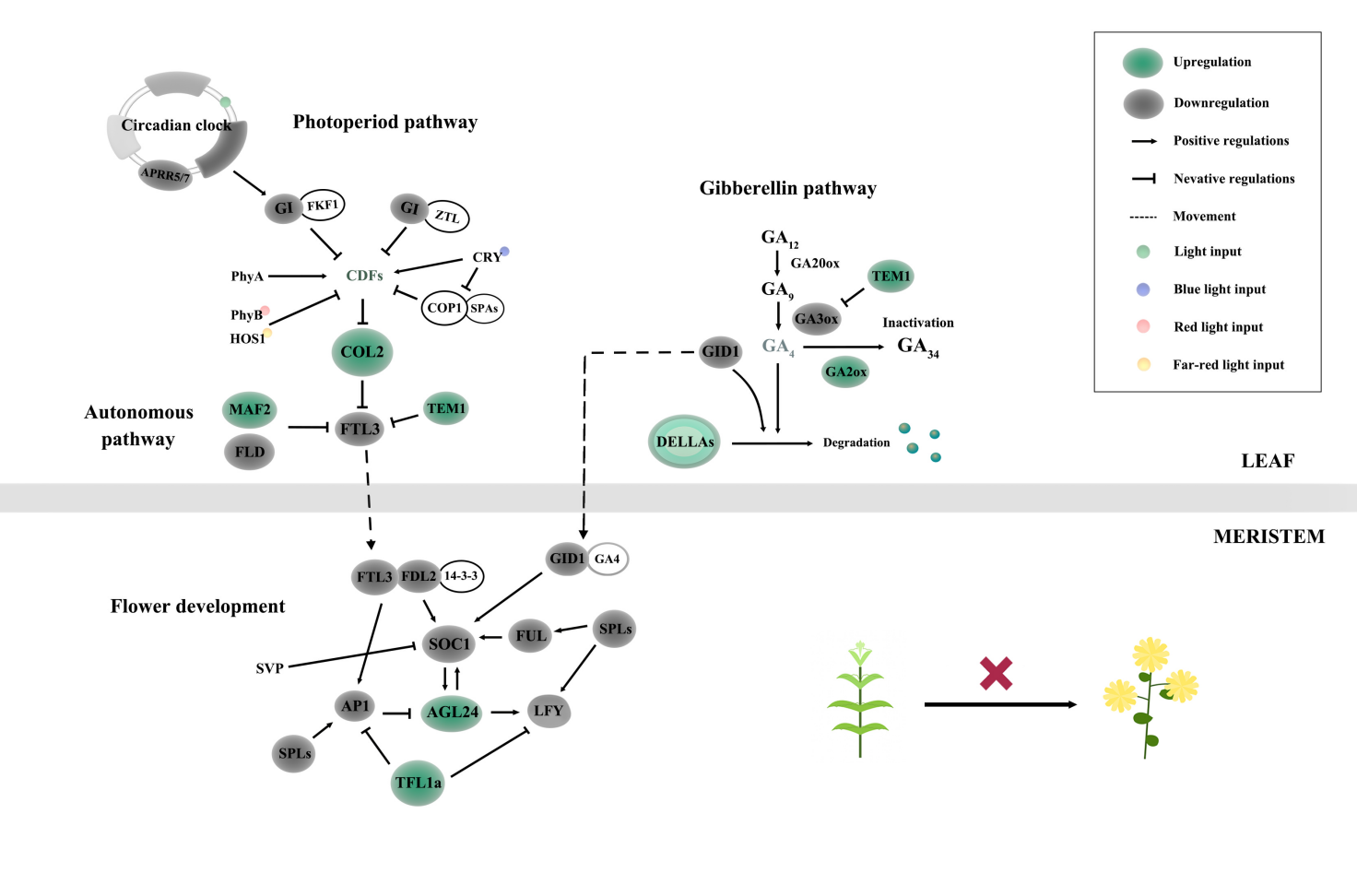
Disrupted regulatory network of flowering in the bud sport of
chrysanthemum ‘Jinshan’.
LEI Ya-Ting
,
FANG Wei-Min
,
CHEN Su-Mei
,
CHEN Fa-Ti
,
JIANG Jia-Fu
.
/
| 〈 |
|
〉 |