

Chinese Bulletin of Botany ›› 2016, Vol. 51 ›› Issue (4): 416-472.DOI: 10.11983/CBB16160 cstr: 32102.14.CBB16160
• REVIEW BY EDITOR-IN-CHIEF • Previous Articles Next Articles
Shuhua Yang1, Tai Wang2, Qian Qian3, Xiaojing Wang4, Jianru Zuo5, Hongya Gu6, Liwen Jiang7, Zhiduan Chen2, Yongfei Bai2, Hongzhi Kong2, Fan Chen5, Langtao Xiao8, Aiwu Dong9, Kang Chong2*
Online:2016-07-01
Published:2016-08-05
About author:# Co-first authors
Shuhua Yang, Tai Wang, Qian Qian, Xiaojing Wang, Jianru Zuo, Hongya Gu, Liwen Jiang, Zhiduan Chen, Yongfei Bai, Hongzhi Kong, Fan Chen, Langtao Xiao, Aiwu Dong, Kang Chong. Research Advances on Plant Science in China in 2015[J]. Chinese Bulletin of Botany, 2016, 51(4): 416-472.
| 国家 | 文章数量 | 所占比例(%) |
|---|---|---|
| 美国 | 367 | 36.6 |
| 中国 | 242 | 24.2 |
| 德国 | 185 | 18.5 |
| 日本 | 111 | 11.1 |
| 法国 | 107 | 10.7 |
Table 1 The number of articles published in The Plant Cell, Plant Physiology, and The Plant Journal from top 5 countries in 2015 (data sources: Web of Science)
| 国家 | 文章数量 | 所占比例(%) |
|---|---|---|
| 美国 | 367 | 36.6 |
| 中国 | 242 | 24.2 |
| 德国 | 185 | 18.5 |
| 日本 | 111 | 11.1 |
| 法国 | 107 | 10.7 |
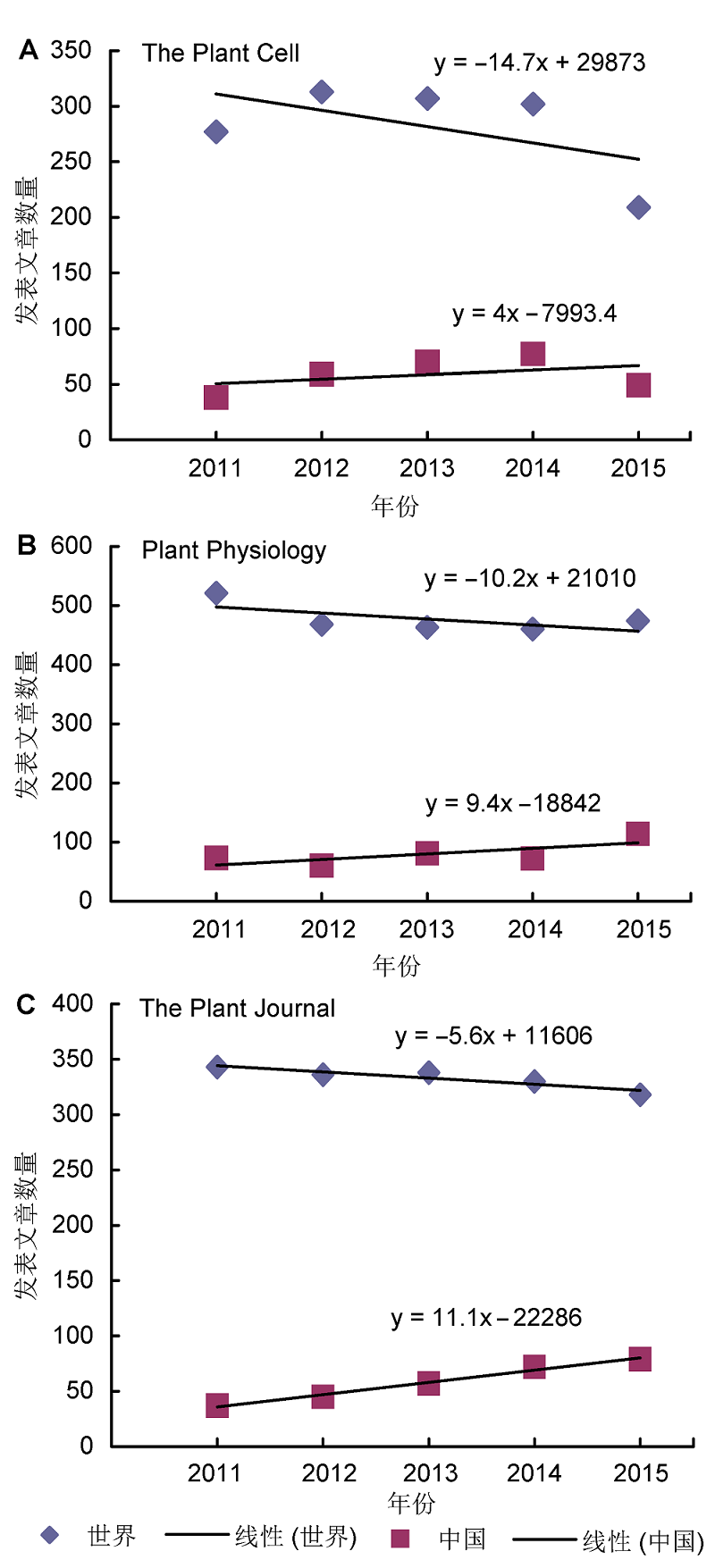
Figure 1 Annual number of plant science publications origi- nating from China and other countries in the world from 2011 to 2015, based on 3 top plant science journals (data sources: Web of Science)
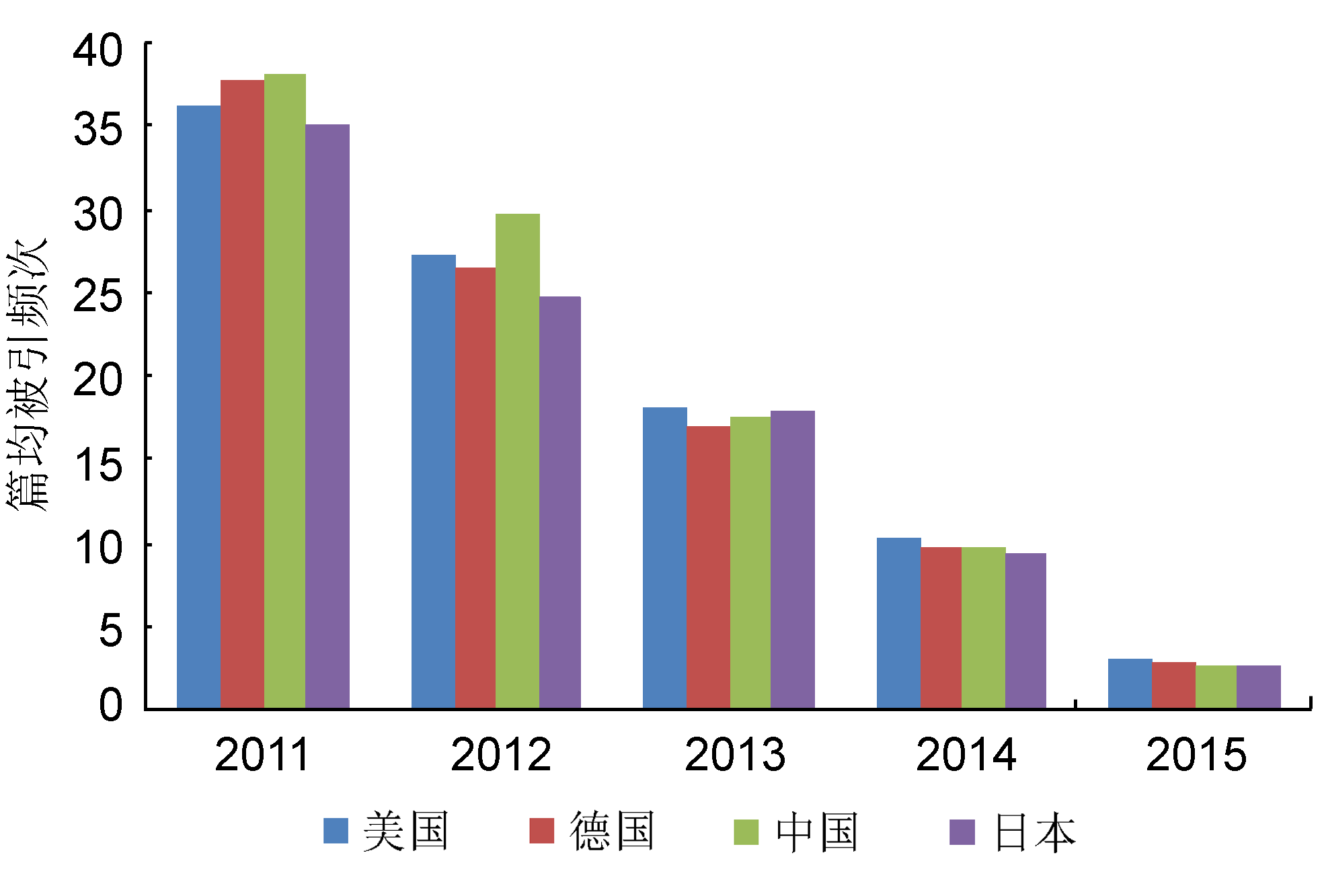
Figure 2 Average citations per paper in China, USA, Germany and Japan during 2011 to 2015, based on 3 top plant science journals (The Plant Cell, Plant Physiology and The Plant Journal) (data sources: Web of Science)
| 1 | 种康, 许智宏, 李家洋 (2016). 中国植物科学基础研究概览. 中国基础科学 (1), 7-12. |
| 2 | 冷静, 王文国, 刘文娟, 李为民, 杜全生, 温明章 (2016). 从国家自然科学基金项目资助看植物科学态势. 植物学报 51, 369-376. |
| 3 | 刘玲童, 王台 (2016). miR396-GRF模块: 水稻分子育种的新资源. 植物学报 51, 148-151. |
| 4 | 施怡婷, 杨淑华 (2016). 中国科学家在乙烯信号转导领域取得突破性进展. 植物学报 51, 287-289. |
| 5 | 许智宏, 种康 (2015). 植物细胞分化与器官发生. 北京: 科学出版社. pp.194-216. |
| 6 | 于倩倩, 孔祥培, 丁兆军 (2015). 中国科学家在生长素信号转导领域取得突破性研究进展. 植物学报 50, 535-537. |
| 7 | 左建儒, 陈凡 (2015). 中国科学家在植物应答低温信号研究中取得突破性进展. 植物学报 50, 145-147. |
| 8 | Adler EM (2016). 2015: signaling breakthroughs of the year. Sci Signal 9, eg1. |
| 9 | Bai J, Zhu X, Wang Q, Zhang J, Chen H, Dong G, Zhu L, Zheng H, Xie Q, Nian J, Chen F, Fu Y, Qian Q, Zuo J (2015). Rice TUTOU1 encodes a suppressor of cAMP receptor-like protein that is important for actin organiza- tion and panicle development.Plant Physiol 169, 1179-1191. |
| 10 | Cai J, Liu X, Vanneste K, Proost S, Tsai WC, Liu KW, Chen LJ, He Y, Xu Q, Bian C, Zheng Z, Sun F, Liu W, Hsiao YY, Pan ZJ, Hsu CC, Yang YP, Hsu YC, Chuang YC, Dievart A, Dufayard JF, Xu X, Wang JY, Wang J, Xiao XJ, Zhao XM, Du R, Zhang GQ, Wang M, Su YY, Xie GC, Liu GH, Li LQ, Huang LQ, Luo YB, Chen HH, Van de Peer Y, Liu ZJ (2015). The genome sequence of the orchid Phalaenopsis equestris.Nat Genet 47, 65-72. |
| 11 | Cao H, Li X, Wang Z, Ding M, Sun Y, Dong F, Chen F, Liu L, Doughty J, Li Y, Liu YX (2015). Histone H2B monou- biquitination mediated by HISTONE MONOUBIQUITINA- TION1 and HISTONE MONOUBIQUITINATION2 is in- volved in anther development by regulating tapetum de- gradation-related genes in rice.Plant Physiol 168, 1389-1405. |
| 12 | Chen HY, Huh JH, Yu YC, Ho LH, Chen LQ, Tholl D, Frommer WB, Guo WJ (2015a). The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon seque- stration from roots and restricts Pythium infection.Plant J 83, 1046-1058. |
| 13 | Chen J, Gao H, Zheng XM, Jin M, Weng JF, Ma J, Ren Y, Zhou K, Wang Q, Wang J, Wang JL, Zhang X, Cheng Z, Wu C, Wang H, Wan JM (2015b). An evolutionarily conserved gene, FUWA, plays a role in determining pani- cle architecture, grain shape and grain weight in rice.Plant J 83, 427-438. |
| 14 | Chen J, Wang Y, Wang F, Yang J, Gao M, Li C, Liu Y, Liu Y, Yamaji N, Ma JF, Paz-Ares J, Nussaume L, Zhang S, Yi K, Wu Z, Wu P (2015c). The rice CK2 kinase regulates tra- fficking of phosphate transporters in response to phosphate levels.Plant Cell 27, 711-723. |
| 15 | Chen J, Yang L, Gu J, Bai X, Ren Y, Fan T, Han Y, Jiang L, Xiao F, Liu Y, Cao S (2015d). MAN3 gene regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis thaliana.New Phytol 205, 570-582. |
| 16 | Chen LY, Grimm GW, Wang QF, Renner SS (2015e). A phylogeny and biogeographic analysis for the capepond- weed family Aponogetonaceae (Alismatales). Mol Phylo- genet Evol 82 Pt A, 111-117. |
| 17 | Chen LY, Shi DQ, Zhang WJ, Tang ZS, Liu J, Yang WC (2015f). The Arabidopsis alkaline ceramidase TOD1 is a key turgor pressure regulator in plant cells.Nat Commun 6, 6030. |
| 18 | Chen WH, Li PF, Chen MK, Lee YI, Yang CH (2015g). FOREVER YOUNG FLOWER negatively regulates ethy- lene response DNA-binding factors by activating an ethy- lene-responsive factor to control Arabidopsis floral organ senescence and abscission.Plant Physiol 168, 1666-1683. |
| 19 | Chen X, Pu H, Fang Y, Wang X, Zhao S, Lin Y, Zhang M, Dai HE, Gong W, Liu L (2015h). Crystal structure of the catalytic subunit of magnesium chelatase.Nat Plants 1, 15125. |
| 20 | Cheng H, Liu H, Deng Y, Xiao J, Li X, Wang S (2015). The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen.Plant Physiol 167, 1087-1099. |
| 21 | Chong K, Xu ZH (2014). Investment in plant research and development bears fruit in China.Plant Cell Rep 33, 541-550. |
| 22 | Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2015). DCA1 acts as a transcriptional co-activator of DST and con- tributes to drought and salt tolerance in rice.PLoS Genet 11, e1005617. |
| 23 | Deng S, Sun J, Zhao R, Ding M, Zhang Y, Sun Y, Wang W, Tan Y, Liu D, Ma X, Hou P, Wang M, Lu C, Shen X, Chen S (2015). Populus euphratica APYRASE2 en- hances cold tolerance by modulating vesicular trafficking and extracellular ATP in Arabidopsis plants.Plant Physiol 169, 530-548. |
| 24 | Ding L, Yan S, Jiang L, Zhao W, Ning K, Zhao J, Liu X, Zhang J, Wang Q, Zhang X (2015a). HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in Arabidopsis.PLoS Genet 11, e1005479. |
| 25 | Ding S, Zhang B, Qin F (2015b). Arabidopsis RZFP34/ CHYR1, a ubiquitin E3 ligase, regulates dtomatal move- ment and frought tolerance via SnRK2.6-mediated phos- phorylation.Plant Cell 27, 3228-3244. |
| 26 | Ding Y, Chang J, Ma Q, Chen L, Liu S, Jin S, Han J, Xu R, Zhu A, Guo J, Luo Y, Xu J, Xu Q, Zeng Y, Deng X, Cheng Y (2015c). Network analysis of postharvest sen- escence process in citrus fruits revealed by transcriptomic and metabolomic profiling.Plant Physiol 168, 357-376. |
| 27 | Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015d). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis.Dev Cell 32, 278-289. |
| 28 | Du SH, Wang ZS, Ingvarsson P, Wang DS, Wang JH, Wu ZQ, Tembrock LR, Zhang JG (2015). Multilocus analysis of nucleotide variation and speciation in three closely related Populus (Salicaceae) species.Mol Ecol 24, 4994-5005. |
| 29 | Duan CG, Wang X, Tang K, Zhang H, Mangrauthia SK, Lei M, Hsu CC, Hou YJ, Wang C, Li Y, Tao WA, Zhu JK (2015). MET18 connects the cytosolic iron-sulfur cluster assembly pathway to active DNA demethylation in Ara- bidopsis.PLoS Genet 11, e1005559. |
| 30 | Fan W, Lou HQ, Gong YL, Liu MY, Cao MJ, Liu Y, Yang JL, Zheng SJ (2015a). Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms.New Phytol 208, 456-468. |
| 31 | Fan XY, Zhang J, Li WJ, Peng LW (2015b). The NdhV subunit is required to stabilize the chloroplast NADH dehydrogenase-like complex in Arabidopsis.Plant J 82, 221-231. |
| 32 | Fang X, Cui Y, Li Y, Qi Y (2015a). Transcription and pro- cessing of primary microRNAs are coupled by elongator complex in Arabidopsis.Nat Plants 1, 15075. |
| 33 | Fang Y, Koba K, Makabe A, Takahashi C, Zhu W, Hayashi T, Hokari AA, Urakawa R, Bai E, Houlton BZ, Xi D, Zhang S, Matsushita K, Tu Y, Liu D, Zhu F, Wang Z, Zhou G, Chen D, Makita T, Toda H, Liu X, Chen Q, Zhang D, Li Y, Yoh M (2015b). Microbial denitrification dominates nitrate losses from forest ecosystems.Proc Natl Acad Sci USA 112, 1470-1474. |
| 34 | Fu YSH, Zhao HF, Piao SL, Peaucelle M, Peng SS, Zhou GY, Ciais P, Huang MT, Menzel A, Peñuelas J, Song Y, Vitasse Y, Zeng ZZ, Janssens IA (2015). Declining global warming effects on the phenology of spring leaf unfolding.Nature 526, 104-107. |
| 35 | Gao F, Wang K, Liu Y, Chen YP, Chen PA, Shi ZY, Luo J, Jiang DQ, Fan FF, Zhu YG, Li SQ (2016). Blocking miR396 increases rice yield by shaping inflorescence architecture.Nat Plants 2, 15196. |
| 36 | Gao J, Wang X, Zhang M, Bian MD, Deng WX, Zuo ZC, Yang ZM, Zhong DP, Lin CT (2015a). Trp triad-de- pendent rapid photoreduction is not required for the function of Arabidopsis CRY1.Proc Natl Acad Sci USA 112, 9135-9140. |
| 37 | Gao XQ, Zhou J, Li J, Zou XW, Zhao JH, Li QL, Xia R, Yang RF, Wang DK, Zuo ZX, Tu J, Tao YZ, Chen XY, Xie Q, Zhu ZR, Qu SH (2015b). Efficient generation of marker-free transgenic rice plants using an improved transposon-mediated transgene reintegration strategy.Plant Physiol 167, 11-24. |
| 38 | Gao YD, Harris AJ, He XJ (2015c). Morphological and ecological divergence of Lilium and Nomocharis within the Hengduan Mountains and Qinghai-Tibetan Plateau may result from habitat specialization and hybridization.BMC Evol Biol 15, 147. |
| 39 | Gardener C, Kumar SV (2015). Hot n' Cold: molecular signatures of domestication bring fresh insights into environmental adaptation.Mol Plant 8, 1439-1441. |
| 40 | Ge XM, Cai HL, Lei X, Zhou X, Yue M, He JM (2015). Heterotrimeric G protein mediates ethylene-induced sto- matal closure via hydrogen peroxide synthesis in Arabi- dopsis.Plant J 82, 138-150. |
| 41 | Geng C, Cong QQ, Li XD, Mou AL, Gao R, Liu JL, Tian YP (2015). DEVELOPMENTALLY REGULATED PLASMA MEMBRANE PROTEIN of Nicotiana benthamiana contri- butes to potyvirus movement and transports to plasmo- desmata via the early secretory pathway and the acto- myosin system.Plant Physiol 167, 394-423. |
| 42 | Gough NR (2015). Rice that tolerates a chill. Sci Signal 8, ec76. |
| 43 | Guan R, Su J, Meng X, Li S, Liu Y, Xu J, Zhang S (2015). Multilayered regulation of ethylene induction plays a posi- tive role in Arabidopsis resistance against Pseudomonas syringae.Plant Physiol 169, 299-312. |
| 44 | Guo DS, Zhang JZ, Wang XL, Han X, Wei BY, Wang JQ, Li BX, Yu H, Huang QP, Gu HY, Qu LJ, Qin GJ (2015a). The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis.Plant Cell 27, 3112-3127. |
| 45 | Guo J, Liu R, Huang L, Zheng XM, Liu PL, Du YS, Cai Z, Zhou L, Wei XH, Zhang FM, Ge S (2016). Widespread and adaptive alterations in genome-wide gene expression associated with ecological divergence of two Oryza spe- cies.Mol Biol Evol 33, 62-78. |
| 46 | Guo LB, Qiu J, Han ZJ, Ye ZH, Chen C, Liu CJ, Xin XF, Ye CY, Wang YY, Xie HQ, Wang Y, Bao JD, Tang S, Xu J, Gui YJ, Fu F, Wang WD, Zhang XC, Zhu QH, Guang XM, Wang CZ, Cui HF, Cai DG, Ge S, Tuskan GA, Yang XH, Qian Q, He SY, Wang J, Zhou XP, Fan LJ (2015b). A host plant genome (Zizania latifolia) after a century-long endophyte infection.Plant J 83, 600-609. |
| 47 | Guo W, Yang H, Liu Y, Gao Y, Ni Z, Peng H, Xin M, Hu Z, Sun Q, Yao Y (2015c). The wheat transcription factor TaGAMyb recruits histone acetyltransferase and activates the expression of a high-molecular-weight glutenin sub- unit gene.Plant J 84, 347-359. |
| 48 | Guo XL, Qin QQ, Yan J, Niu YL, Huang BY, Guan LP, Li Y, Ren DT, Li J, Hou SW (2015d). Type-one protein phosphatase4 regulates pavement cell interdigitation by modulating Pin-Formed1 polarity and trafficking in Ara- bidopsis.Plant Physiol 167, 1058-1075. |
| 49 | Han SJ, Wang Y, Zheng XY, Jia Q, Zhao JP, Bai F, Hong YG, Liu YL (2015a). Cytoplastic glyceraldehyde-3-phos- phate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana bentha- miana.Plant Cell 27, 1316-1331. |
| 50 | Han TS, Wu Q, Hou XH, Li ZW, Zou YP, Ge S, Guo YL (2015b). Frequent introgressions from diploid species contribute to the adaptation of the tetraploid Shepherd's purse (Capsella bursa-pastoris).Mol Plant 8, 427-438. |
| 51 | Han Y, Dang R, Li J, Jiang J, Zhang N, Jia M, Wei L, Li Z, Li B, Jia W (2015c). SUCROSE NONFERMENTING1- RELATED PROTEIN KINASE2.6, an ortholog of OPEN STOMATA1, is a negative regulator of strawberry fruit development and ripening.Plant Physiol 167, 915-930. |
| 52 | Hao ZZ, Liu YY, Nazaire M, Wei XX, Wang XQ (2015). Molecular phylogenetics and evolutionary history of sect. Quinquefoliae (Pinus): implications for Northern Hemis- phere biogeography.Mol Phylogenet Evol 87, 65-79. |
| 53 | Hsu CC, Chen YY, Tsai WC, Chen WH, Chen HH (2015a). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp.Plant Physiol 168, 175-191. |
| 54 | Hsu HF, Hsu WH, Lee YI, Mao WT, Yang JY, Li JY, Yang CH (2015b). Model for perianth formation in orchids.Nat Plants 1, 15046. |
| 55 | Hu B, Wang W, Ou SJ, Tang JY, Li H, Che RH, Zhang ZH, Chai XY, Wang HR, Wang YQ, Liang CZ, Liu LC, Piao ZZ, Deng QY, Deng K, Xu C, Liang Y, Zhang LH, Li LG, Chu CC (2015a). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies.Nat Ge- net 47, 834-838. |
| 56 | Hu FH, Zhu Y, Wu WJ, Xie Y, Huang JR (2015b). Leaf variegation of Thylakoid Formation1 is suppressed by mutations of specific sigma-factors in Arabidopsis.Plant Physiol 168, 1066-1075. |
| 57 | Hu L, Ye M, Li R, Zhang T, Zhou G, Wang Q, Lu J, Lou Y (2015c). The rice transcription factor WRKY53 supp- resses herbivore-induced defenses by acting as a neg- ative feedback modulator of mitogen-activated protein kinase activity.Plant Physiol 169, 2907-2921. |
| 58 | Hu Z, Song N, Zheng M, Liu X, Liu Z, Xing J, Ma J, Guo W, Yao Y, Peng H, Xin M, Zhou DX, Ni Z, Sun Q (2015d). Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotoler- ance in Arabidopsis.Plant J 84, 1178-1191. |
| 59 | Hua L, Wang DR, Tan LB, Fu YC, Liu FX, Xiao LT, Zhu ZF, Fu Q, Sun XY, Gu P, Cai HW, McCouch SR, Sun CQ (2015). LABA1, a domestication gene associated with long, barbed awns in wild rice.Plant Cell 27, 1875-1888. |
| 60 | Huang CH, Sun RR, Hu Y, Zeng LP, Zhang N, Cai LM, Zhang Q, Koch MA, Al-Shehbaz I, Edger PP, Pires JC, Tan DY, Zhong Y, Ma H (2016). Resolution of Brassi- caceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolu- tion.Mol Biol Evol 33, 394-412. |
| 61 | Huang DB, Wang SG, Zhang BC, Shang-Guan KK, Shi YY, Zhang DM, Liu XL, Wu K, Xu ZP, Fu XD, Zhou YH (2015a). A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice.Plant Cell 27, 1681-1696. |
| 62 | Huang MK, Hu YL, Liu X, Li YG, Hou XL (2015b). Arabidopsis LEAFY COTYLEDON1 mediates postembr- yonic development via interacting with PHYTOCHROME- INTERACTING FACTOR4.Plant Cell 27, 3099-3111. |
| 63 | Huang W, Yu C, Hu J, Wang L, Dan Z, Zhou W, He C, Zeng Y, Yao G, Qi J, Zhang Z, Zhu R, Chen X, Zhu Y (2015c). Pentatricopeptide-repeat family protein RF6 func- tions with hexokinase 6 to rescue rice cytoplasmic male sterility.Proc Natl Acad Sci USA 112, 14984-14989. |
| 64 | Huang X, Yang S, Gong J, Zhao Y, Feng Q, Gong H, Li W, Zhan Q, Cheng B, Xia J, Chen N, Hao Z, Liu K, Zhu C, Huang T, Zhao Q, Zhang L, Fan D, Zhou C, Lu Y, Weng Q, Wang ZX, Li J, Han B (2015d). Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis.Nat Commun 6, 6258. |
| 65 | Ji H, Wang Y, Cloix C, Li K, Jenkins GI, Wang S, Shang Z, Shi Y, Yang S, Li X (2015a). The Arabidopsis RCC1 family protein TCF1 regulates freezing tolerance and cold acclimation through modulating lignin biosynthesis.PLoS Genet 11, e1005471. |
| 66 | Ji X, Zhang H, Zhang Y, Wang Y, Gao C (2015b). Est- ablishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants.Nat Plants 1, 15144. |
| 67 | Jiang J, Zhang C, Wang X (2015). A recently evolved isoform of the transcription factor BES1 promotes brass- inosteroid signaling and development in Arabidopsis tha- liana.Plant Cell 27, 361-374. |
| 68 | Jin J, Shi J, Liu B, Liu Y, Huang Y, Yu Y, Dong A (2015a). MORF-RELATED GENE702, a reader protein of trime- thylated histone H3 lysine 4 and histone H3 lysine 36, is involved in brassinosteroid-regulated growth and flow- ering time control in rice.Plant Physiol 168, 1275-1285. |
| 69 | Jin JP, He K, Tang X, Li Z, Lv L, Zhao Y, Luo JC, Gao G (2015b). An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors.Mol Biol Evol 32, 1767-1773. |
| 70 | Jing H, Yang X, Zhang J, Liu X, Zheng H, Dong G, Nian J, Feng J, Xia B, Qian Q, Li J, Zuo J (2015a). Pep- tidyl-prolyl isomerization targets rice Aux/IAAs for prote- asomal degradation during auxin signaling.Nat Commun 6, 7395. |
| 71 | Jing X, Sanders NJ, Shi Y, Chu H, Classen AT, Zhao K, Chen L, Shi Y, Jiang Y, He J (2015b). The links between ecosystem multifunctionality and above- and belowground biodiversity are mediated by climate.Nat Commun 6, 8159. |
| 72 | Kim C, Deng T, Wen J, Nie ZL, Sun H (2015). Systematics, biogeography, and character evolution of Deutzia (Hy- drangeaceae) inferred from nuclear and chloroplast DNA sequences.Mol Phylogenet Evol 87, 91-104. |
| 73 | Kong L, Cheng J, Zhu Y, Ding Y, Meng J, Chen Z, Xie Q, Guo Y, Li J, Yang S, Gong Z (2015). Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases.Nat Commun 6, 8630. |
| 74 | Lan ZC, Jenerette GD, Zhan SX, Li WH, Zheng SX, Bai YF (2015). Testing the scaling effects and mechanisms of N-induced biodiversity loss: evidence from a decade-long grassland experiment.J Ecol 103, 750-760. |
| 75 | Law YS, Zhang RS, Guan XQ, Cheng SF, Sun F, Duncan O, Murcha MW, Whelan J, Lim BL (2015). Phos- phorylation and dephosphorylation of the presequence of precursor MULTIPLE ORGANELLAR RNA EDITING FACTOR3 during import into mitochondria from Arabi- dopsis.Plant Physiol 169, 1344-1355. |
| 76 | Lee DH, Kao YH, Ku JC, Lin CY, Meeley R, Jan YS, Wang CJR (2015). The axial element protein DESYNAPTIC2 mediates meiotic double-strand break formation and synaptonemal complex assembly in maize.Plant Cell 27, 2516-2529. |
| 77 | Lei L, Shi J, Chen J, Zhang M, Sun S, Xie S, Li X, Zeng B, Peng L, Hauck A, Zhao H, Song W, Fan Z, Lai J (2015a). Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress.Plant J 84, 1206-1218. |
| 78 | Lei M, Zhang H, Julian R, Tang K, Xie S, Zhu J (2015b). Regulatory link between DNA methylation and active demethylation in Arabidopsis.Proc Natl Acad Sci USA 112, 3553-3557. |
| 79 | Lei MJ, Wang Q, Li XL, Chen AM, Luo L, Xie YJ, Li G, Luo D, Mysore KS, Wen JQ, Xie ZP, Staehelin C, Wang YZ (2015c). The small GTPase ROP10 of Medicago trun- catula is required for both tip growth of root hairs and nod factor-induced root hair deformation.Plant Cell 27, 806-822. |
| 80 | Li C, Qiao Z, Qi W, Wang Q, Yuan Y, Yang X, Tang Y, Mei B, Lv Y, Zhao H, Xiao H, Song R (2015a). Genome-wide characterization of cis-acting DNA targets reveals the tr- anscriptional regulatory framework of opaque2 in maize.Plant Cell 27, 532-545. |
| 81 | Li HJ, Zhu SS, Zhang MX, Wang T, Liang L, Xue Y, Shi DQ, Liu J, Yang WC (2015b). Arabidopsis CBP1 is a novel regulator of transcription initiation in central cell- mediated pollen tube guidance.Plant Cell 27, 2880-2893. |
| 82 | Li J, Liu JT, Wang GQ, Cha JY, Li GN, Chen S, Li Z, Guo JH, Zhang CG, Yang YQ, Kim WY, Yun DJ, Schumaker KS, Chen ZZ, Guo Y (2015c). A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis.Plant Cell 27, 908-925. |
| 83 | Li JT, Zhao Y, Chu HW, Wang LK, Fu YR, Liu P, Upadhyaya N, Chen CL, Mou TM, Feng YQ, Kumar P, Xu J (2015d). SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation.PLoS Genet 11, e1005464. |
| 84 | Li MR, Shi FX, Zhou YX, Li YL, Wang XF, Zhang C, Wang XT, Liu B, Xiao HX, Li LF (2015e). Genetic and epi- genetic diversities shed light on domestication of culti- vated ginseng (Panax ginseng).Mol Plant 8, 1612-1622. |
| 85 | Li Q, Wang X, Sun H, Zeng J, Cao Z, Li Y, Qian W (2015f). Regulation of active DNA demethylation by a methyl- CpG-binding domain protein in Arabidopsis thaliana.PLoS Genet 11, e1005210. |
| 86 | Li R, Zhu S, Chen HYH, John R, Zhou G, Zhang D, Zhang Q, Ye Q (2015g). Are functional traits a good predictor of global change impacts on tree species abundance dyna- mics in a subtropical forest?Ecol Lett 18, 1181-1189. |
| 87 | Li S, Wang H, Li F, Chen Z, Li X, Zhu L, Wang G, Yu J, Huang D, Lang Z (2015h). The maize transcription factor EREB58 mediates the jasmonate-induced production of sesquiterpene volatiles.Plant J 84, 296-308. |
| 88 | Li SP, Cadotte MW, Meiners SJ, Hua ZS, Shu HY, Li JT, Shu WS (2015i). The effects of phylogenetic relatedness on invasion success and impact: deconstructing Darwin's natura- lisation conundrum.Ecol Lett 18, 1285-1292. |
| 89 | Li WY, Ma MD, Feng Y, Li HJ, Wang YC, Ma YT, Li MZ, An FY, Guo HW (2015j). EIN2-directed translational regu- lation of ethylene signaling in Arabidopsis.Cell 163, 670-683. |
| 90 | Li X, Li L, Yan J (2015k). Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize.Nat Commun 6, 6648. |
| 91 | Li X, Lin HX (2016). Molecular signature of chilling adap- tation in rice. Natl Sci Rev 3(3). (in Press) |
| 92 | Li XM, Chao DY, Wu Y, Huang X, Chen K, Cui LG, Su L, Ye WW, Chen H, Chen HC, Dong NQ, Guo T, Shi M, Feng Q, Zhang P, Han B, Shan JX, Gao JP, Lin HX (2015l). Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice.Nat Genet 47, 827-833. |
| 93 | Lin L, Tang L, Bai YJ, Tang ZY, Wang W, Chen ZD (2015a). Range expansion and habitat shift triggered elevated diversification of the rice genus (Oryza, Poac- eae) during the Pleistocene.BMC Evol Biol 15, 182. |
| 94 | Lin Q, Ohashi Y, Kato M, Tsuge T, Gu H, Qu L, Aoyama T (2015b). GLABRA2 directly suppresses basic helix-loop- helix transcription factor genes with diverse functions in root hair development.Plant Cell 27, 2894-2906. |
| 95 | Lin Q, Wu F, Sheng P, Zhang Z, Zhang X, Guo X, Wang J, Cheng Z, Wang J, Wang H, Wan J (2015c). The SnRK2-APC/C (TE) regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways.Nat Commun 6, 7981. |
| 96 | Lin Y, Wong WO, Shi G, Shen S, Li Z (2015d). Bilobate leaves of bauhinia (Leguminosae, Caesalpinioideae, Cer- cideae) from the middle Miocene of Fujian province, sou- theastern China and their biogeographic implications.BMC Evol Biol 15, 252. |
| 97 | Lin YS, Ding Y, Wang J, Shen JB, Kung CH, Zhuang XH, Cui Y, Yin Z, Xia YJ, Lin HX, Robinson DG, Jiang LW (2015e). Exocyst-positive organelles and autophagosom- es are distinct organelles in plants.Plant Physiol 169, 1917-1932. |
| 98 | Liu HH, Liu HQ, Zhou LN, Zhang ZH, Zhang X, Wang ML, Li HX, Lin ZW (2015a). Parallel domestication of the Heading Date 1 gene in cereals.Mol Biol Evol 32, 2726-2737. |
| 99 | Liu HJ, Tang ZX, Han XM, Yang ZL, Zhang FM, Yang HL, Liu YJ, Zeng QY (2015b). Divergence in enzymatic activities in the soybean GST supergene family provides new insight into the evolutionary dynamics of wholegeno- me duplicates.Mol Biol Evol 32, 2844-2859. |
| 100 | Liu J, Hua W, Hu ZY, Yang HL, Zhang L, Li RJ, Deng LB, Sun XC, Wang XF, Wang HZ (2015c). Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed.Proc Natl Acad Sci USA 112, E5123-E5132. |
| 101 | Liu J, Yang H, Bao F, Ao K, Zhang X, Zhang Y, Yang S (2015d). IBR5 modulates temperature-dependent, R pro- tein CHS3-mediated defense responses in Arabidopsis.PLoS Genet 11, e1005584. |
| 102 | Liu JL, Yang L, Luan MD, Wang Y, Zhang C, Zhang B, Shi JS, Zhao FG, Lan WZ, Luan S (2015e). A vacuolar phosphate transporter essential for phosphate homeosta- sis in Arabidopsis.Proc Natl Acad Sci USA 112, E6571-E6578. |
| 103 | Liu JW, Zhou WB, Liu GF, Yang CP, Sun Y, Wu WJ, Cao SQ, Wang C, Hai GH, Wang ZF, Bock R, Huang JR, Cheng YX (2015f). The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA proces- sing in Arabidopsis.Plant Physiol 168, 205-221. |
| 104 | Liu LC, Tong HN, Xiao YH, Che RH, Xu F, Hu B, Liang CZ, Chu JF, Li JY, Chu CC (2015g). Activation of Big Grain1 significantly improves grain size by regulating auxin trans- port in rice.Proc Natl Acad Sci USA 112, 11102-11107. |
| 105 | Liu R, Zheng XM, Zhou L, Zhou HF, Ge S (2015h). Population genetic structure of Oryza rufipogon and Oryza nivara: implications for the origin of O. nivara.Mol Ecol 24, 5211-5228. |
| 106 | Liu X, Zhou S, Wang W, Ye Y, Zhao Y, Xu Q, Zhou C, Tan F, Cheng S, Zhou D (2015i). Regulation of histone methylation and reprogramming of gene expression in the rice inflorescence meristem.Plant Cell 27, 1428-1444. |
| 107 | Liu YD, Zhang CC, Wang DH, Su W, Liu LC, Wang MY, Li JM (2015j). EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated de- gradation system in Arabidopsis.Proc Natl Acad Sci USA 112, 12205-12210. |
| 108 | Liu Z, Yan JP, Li DK, Luo Q, Yan Q, Liu ZB, Ye LM, Wang JM, Li XF, Yang Y (2015k). UDP-glucosyltransferase- 71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis.Plant Physiol 167, 1659-1670. |
| 109 | Luo M, Tai R, Yu CW, Yang S, Chen CY, Lin WD, Schmidt W, Wu K (2015). Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis.Plant J 82, 925-936. |
| 110 | Lyu MJ, Gowik U, Kelly S, Covshoff S, Mallmann J, Westhoff P, Hibberd JM, Stata M, Sage RF, Lu H, Wei X, Wong GK, Zhu XG (2015). RNA-Seq based phylogeny recapitulates previous phylogeny of the genus Flaveria (Asteraceae) with some modifications.BMC Evol Biol 15, 116. |
| 111 | Ma J, Cai H, He C, Zhang W, Wang L (2015a). A hemi- cellulose-bound form of silicon inhibits cadmium ion up- take in rice (Oryza sativa) cells.New Phytol 206, 1063-1074. |
| 112 | Ma M, Wang Q, Li Z, Cheng H, Li Z, Liu X, Song W, Appels R, Zhao H (2015b). Expression of TaCYP78A3, a gene encoding cytochrome P450 CYP78A3 protein in wheat (Triticum aestivum L.), affects seed size.Plant J 83, 312-325. |
| 113 | Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, Xiao J, Guo X, Xu S, Niu Y, Jin J, Zhang H, Xu X, Li L, Wang W, Qian Q, Ge S, Chong K (2015c). COLD1 confers chilling tolerance in rice.Cell 160, 1209-1221. |
| 114 | Ma Z, Song T, Zhu L, Ye W, Wang Y, Shao Y, Dong S, Zhang Z, Dou D, Zheng X, Tyler BM (2015d). A phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP.Plant Cell 27, 2057-2072. |
| 115 | Manishankar P, Kudla J (2015). Cold tolerance encoded in one SNP.Cell 160, 1045-1046. |
| 116 | Mao H, Wang H, Liu S, Li Z, Yang X, Yan J, Li J, Tran LS, Qin F (2015a). A transposable element in a NAC gene is associated with drought tolerance in maize seedlings.Nat Commun 6, 8326. |
| 117 | Mao J, Chi W, Ouyang M, He BY, Chen F, Zhang LX (2015b). PAB is an assembly chaperone that functions downstream of chaperonin 60 in the assembly of chlo- roplast ATP synthase coupling factor 1.Proc Natl Acad Sci USA 112, 4152-4157. |
| 118 | Merchante C, Brumos J, Yun J, Hu QW, Spencer KR, Enriquez P, Binder BM, Heber S, Stepanova AN, Alonso JM (2015). Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2.Cell 163, 684-697. |
| 119 | Min L, Hu Q, Li Y, Xu J, Ma Y, Zhu L, Yang X, Zhang X (2015). LEAFY COTYLEDON1-CASEIN KINASE I-TCP- 15-PHYTOCHROME INTERACTING FACTOR4 network regulates somatic embryogenesis by regulating auxin ho- meostasis.Plant Physiol 169, 2805-2821. |
| 120 | Ming R, VanBuren R, Wai CM, Tang HB, Schatz MC, Bowers JE, Lyons E, Wang ML, Chen J, Biggers E, Zhang JS, Huang LX, Zhang LM, Miao WJ, Zhang J, Ye ZY, Miao CY, Lin ZC, Wang H, Zhou HY, Yim WC, Priest HD, Zheng CF, Woodhouse M, Edger PP, Guyot R, Guo HB, Guo H, Zheng GY, Singh R, Sharma A, Min XJ, Zheng Y, Lee H, Gurtowski J, Sedlazeck FJ, Harkess A, McKain MR, Liao ZY, Fang JP, Liu J, Zhang XD, Zhang Q, Hu WC, Qin Y, Wang K, Chen LY, Shirley N, Lin YR, Liu LY, Hernandez AG, Wright CL, Bulone V, Tuskan GA, Heath K, Zee F, Moore PH, Sunkar R, Leebens-Mack JH, Mockler T, Bennetzen JL, Freeling M, Sankoff D, Paterson AH, Zhu XG, Yang XH, Smith JAC, Cushman JC, Paull RE, Yu QY (2015). The pineapple genome and the evolution of CAM photosynthesis.Nat Genet 47, 1435-1442. |
| 121 | Nie ZL, Funk VA, Meng Y, Deng T, Sun H, Wen J (2016). Recent assembly of the global herbaceous flora: evidence from the paper daisies (Asteraceae: Gnaphalieae).New Phytol 209, 1795-1806. |
| 122 | Niu BX, Wang LD, Zhang LS, Ren D, Ren R, Copenhaver GP, Ma H, Wang YX (2015). Arabidopsis cell division cycle 20.1 is required for normal meiotic spindle assembly and chromosome segregation.Plant Cell 27, 3367-3382. |
| 123 | Niu SH, Yuan HW, Sun XR, Porth I, Li Y, El-Kassaby YA, Li W (2016). A transcriptomics investigation into pine reproductive organ development.New Phytol 209, 1278-1289. |
| 124 | Peng Y, Chen L, Lu Y, Wu Y, Dumenil J, Zhu Z, Bevan MW, Li Y (2015). The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modu- lating the stability of TCP14/15 in Arabidopsis.Plant Cell 27, 649-662. |
| 125 | Pikaard CS, Li Y, Córdoba-Cañero D, Qian W, Zhu X, Tang K, Zhang H, Ariza RR, Roldán-Arjona T, Zhu JK (2015). An AP endonuclease functions in active DNA demethylation and gene imprinting in Arabidopsis.PLoS Genet 11, e1004905. |
| 126 | Qi T, Huang H, Song S, Xie D (2015a). Regulation of jasmonate-mediated stamen development and seed pro- duction by a bHLH-MYB complex in Arabidopsis.Plant Cell 27, 1620-1633. |
| 127 | Qi T, Wang J, Huang H, Liu B, Gao H, Liu Y, Song S, Xie D (2015b). Regulation of jasmonate-induced leaf senes- cence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis.Plant Cell 27, 1634-1649. |
| 128 | Qin X, Suga M, Kuang T, Shen JR (2015). Photosynthesis. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex.Science 348, 989-995. |
| 129 | Qiu K, Li Z, Yang Z, Chen J, Wu S, Zhu X, Gao S, Gao J, Ren G, Kuai B, Zhou X (2015). EIN3 and ORE1 acc- elerate degreening during ethylene-mediated leaf sen- escence by directly activating chlorophyll catabolic genes in Arabidopsis.PLoS Genet 11, e1005399. |
| 130 | Ran JH, Shen TT, Liu WJ, Wang PP, Wang XQ (2015). Mitochondrial introgression and complex biogeographic history of the genus Picea.Mol Phylogenet Evol 93, 63-76. |
| 131 | Rao Y, Yang Y, Xu J, Li X, Leng Y, Dai L, Huang L, Shao G, Ren D, Hu J, Guo L, Pan J, Zeng D (2015). EARLY SENESCENCE1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice.Plant Physiol 169, 1225-1239. |
| 132 | Raskoti BB, Jin WT, Xiang XG, Schuiteman A, Li DZ, Li JW, Huang WC, Jin XH, Huang LQ (2016). A phylo- genetic analysis of molecular and morphological charac- ters of Herminium (Orchidaceae, Orchideae): evolutiona- ry relationships, taxonomy, and patterns of character evolution.Cladistics 32, 198-210. |
| 133 | Shi C, Qi C, Ren H, Huang A, Hei S, She X (2015a). Ethylene mediates brassinosteroid-induced stomatal clo- sure via Galpha protein-activated hydrogen peroxide and nitric oxide production in Arabidopsis.Plant J 82, 280-301. |
| 134 | Shi H, Wang X, Mo XR, Tang C, Zhong SW, Deng XW (2015b). Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination.Proc Natl Acad Sci USA 112, 3817-3822. |
| 135 | Shi Y, Gong Z (2015). One SNP in COLD1 determines cold tolerance during rice domestication.J Genet Genomics 42, 133-134. |
| 136 | Shi Y, Yang S (2015). COLD1: a cold sensor in rice.Sci China Life Sci 58, 409-410. |
| 137 | Song ZT, Sun L, Lu SJ, Tian Y, Ding Y, Liu JX (2015). Transcription factor interaction with COMPASS-like com- plex regulates histone H3K4 trimethylation for specific gene expression in plants.Proc Natl Acad Sci USA 112, 2900-2905. |
| 138 | Su T, Xu Q, Zhang FC, Chen Y, Li LQ, Wu WH, Chen YF (2015). WRKY42 modulates phosphate homeostasis thr- ough regulating phosphate translocation and acquisition in Arabidopsis.Plant Physiol 167, 1579-1591. |
| 139 | Sun L, Zhang A, Zhou Z, Zhao Y, Yan A, Bao S, Yu H, Gan Y (2015a). GLABROUS INFLORESCENCE STEM- S3 (GIS3) regulates trichome initiation and development in Arabidopsis.New Phytol 206, 220-230. |
| 140 | Sun LC (2015). A closer mimic of the oxygen evolution complex of photosystem II.Science 348, 635-636. |
| 141 | Sun M, Soltis DE, Soltis PS, Zhu X, Burleigh JG, Chen Z (2015b). Deep phylogenetic incongruence in the angio- sperm clade Rosidae.Mol Phylogenet Evol 83, 156-166. |
| 142 | Sun Y, Xu W, Jia Y, Wang M, Xia G (2015c). The wheat TaGBF1 gene is involved in the blue-light response and salt tolerance.Plant J 84, 1219-1230. |
| 143 | Sun Z, Guo T, Liu Y, Liu Q, Fang Y (2015d). The roles of Arabidopsis CDF2 in transcriptional and posttranscrip- tional regulation of primary microRNAs.PLoS Genet 11, e1005598. |
| 144 | Tang K, Ding WL, Hoppner A, Zhao C, Zhang L, Hontani Y, Kennis JT, Gartner W, Scheer H, Zhou M, Zhao KH (2015a). The terminal phycobilisome emitter, LCM: a light- harvesting pigment with a phytochrome chromophore.Proc Natl Acad Sci USA 112, 15880-15885. |
| 145 | Tang L, Zou XH, Zhang LB, Ge S (2015b). Multilocus species tree analyses resolve the ancient radiation of the subtribe Zizaniinae (Poaceae).Mol Phylogenet Evol 84, 232-239. |
| 146 | Tang RJ, Zhao FG, Garcia VJ, Kleist TJ, Yang L, Zhang HX, Luan S (2015c). Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis.Proc Natl Acad Sci USA 112, 3134-3139. |
| 147 | Tang Y, Yukawa T, Bateman RM, Jiang H, Peng H (2015d). Phylogeny and classification of the East Asian Amitostigma alliance (Orchidaceae: Orchideae) based on six DNA markers.BMC Evol Biol 15, 96. |
| 148 | Tian J, Han LB, Feng ZD, Wang GD, Liu WW, Ma YP, Yu YJ, Kong ZS (2015a). Orchestration of microtubules and the actin cytoskeleton in trichome cell shape determina- tion by a plant-unique kinesin.Elife 4, e09351. |
| 149 | Tian S, Lei SQ, Hu W, Deng LL, Li B, Meng QL, Soltis DE, Soltis PS, Fan DM, Zhang ZY (2015b). Repeated range expansions and inter-/postglacial recolonization routes of Sargentodoxa cuneata (Oliv.) Rehd. et Wils. (Lardizaba- laceae) in subtropical China revealed by chloroplast phy- logeography.Mol Phylogenet Evol 85, 238-246. |
| 150 | Tu B, Liu L, Xu C, Zhai J, Li S, Lopez MA, Zhao Y, Yu Y, Ramachandran V, Ren G, Yu B, Li S, Meyers BC, Mo B, Chen X (2015). Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis.PLoS Genet 11, e1005119. |
| 151 | Wan JM (2015). Engineering thermotolerant plants: a sol- ution to protecting crop production threatened by global warming.Sci Bull 60, 1366-1367. |
| 152 | Wang B, Chu J, Yu T, Xu Q, Sun X, Yuan J, Xiong G, Wang G, Wang Y, Li J (2015a). Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis.Proc Natl Acad Sci USA 112, 4821-4826. |
| 153 | Wang C, Dong X, Jin D, Zhao Y, Xie S, Li X, He X, Lang Z, Lai J, Zhu JK, Gong Z (2015b). Methyl-CpG-binding domain protein MBD7 is required for active DNA deme- thylation in Arabidopsis.Plant Physiol 167, 905-914. |
| 154 | Wang C, Yue WH, Ying YH, Wang SD, Secco D, Liu Y, Whelan J, Tyerman SD, Shou HX (2015c). Rice SPX- major facility superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeos- tasis in rice.Plant Physiol 169, 2822-2831. |
| 155 | Wang C, Zhu MS, Duan LJ, Yu HX, Chang XJ, Li L, Kang H, Feng Y, Zhu H, Hong ZL, Zhang ZM (2015d). Lotus japonicus Clathrin Heavy Chain1 is associated with Rho- Like GTPase ROP6 and involved in nodule formation.Plant Physiol 167, 1497-1510. |
| 156 | Wang F, Chen HW, Li QT, Wei W, Li W, Zhang WK, Ma B, Bi YD, Lai YC, Liu XL, Man WQ, Zhang JS, Chen SY (2015e). GmWRKY27 interacts with GmMYB174 to re- duce expression of GmNAC29 for stress tolerance in soy- bean plants.Plant J 83, 224-236. |
| 157 | Wang HZ, Yang KZ, Zou JJ, Zhu LL, Xie ZD, Morita MT, Tasaka M, Friml J, Grotewold E, Beeckman T, Vanneste S, Sack F, Le J (2015f). Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism.Nat Commun 6, 8822. |
| 158 | Wang J, Yao W, Zhu D, Xie WB, Zhang QF (2015g). Genetic basis of sRNA quantitative variation analyzed using an experimental population derived from an elite rice hybrid.Elife 4, e04250. |
| 159 | Wang JJ, Guo HS (2015). Cleavage of INDOLE-3-ACETIC ACID INDUCIBLE28 mRNA by microRNA847 upregulates auxin signaling to modulate cell proliferation and lateral organ growth in Arabidopsis.Plant Cell 27, 574-590. |
| 160 | Wang JP, Chuang L, Loziuk PL, Chen H, Lin YC, Shi R, Qu GZ, Muddiman DC, Sederoff RR, Chiang VL (2015h). Phosphorylation is an on/off switch for 5-hydro- xyconiferaldehyde O-methyltransferase activity in poplar monolignol biosynthesis.Proc Natl Acad Sci USA 112, 8481-8486. |
| 161 | Wang L, Sun SY, Jin JY, Fu DB, Yang XF, Weng XY, Xu CG, Li XH, Xiao JH, Zhang QF (2015i). Coordinated regulation of vegetative and reproductive branching in rice.Proc Natl Acad Sci USA 112, 15504-15509. |
| 162 | Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J (2015j). Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation.Plant Cell 27, 3128-3142. |
| 163 | Wang M, Yuan D, Tu L, Gao W, He Y, Hu H, Wang P, Liu N, Lindsey K, Zhang X (2015k). Long noncoding RNAs and their proposed functions in fibre development of cot- ton (Gossypium spp.).New Phytol 207, 1181-1197. |
| 164 | Wang P, Du Y, Hou YJ, Zhao Y, Hsu CC, Yuan F, Zhu X, Tao WA, Song CP, Zhu JK (2015l). Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1.Proc Natl Acad Sci USA 112, 613-618. |
| 165 | Wang PP, Liao H, Zhang WG, Yu XX, Zhang R, Shan HY, Duan XS, Yao X, Kong HZ (2016). Flexibility in the structure of spiral flowers and its underlying mechanisms.Nat Plants 2, 15188. |
| 166 | Wang R, Jing W, Xiao L, Jin Y, Shen L, Zhang W (2015m). The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-Type trans- cription factor.Plant Physiol 168, 1076-1090. |
| 167 | Wang SH, Yang XY, Xu MN, Lin XZ, Lin T, Qi JJ, Shao GJ, Tian NN, Yang Q, Zhang ZH, Huang SW (2015n). A rare SNP identified a TCP transcription factor essential for tendril development in cucumber.Mol Plant 8, 1795-1808. |
| 168 | Wang SK, Li S, Liu Q, Wu K, Zhang JQ, Wang SS, Wang Y, Chen XB, Zhang Y, Gao CX, Wang F, Huang HX, Fu XD (2015o). The OsSPL16-GW7 regulatory module de- termines grain shape and simultaneously improves rice yield and grain quality.Nat Genet 47, 949-954. |
| 169 | Wang X, Zhang S, Dou Y, Zhang C, Chen X, Yu B, Ren G (2015p). Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3' tailing of small RNAs in Arabidopsis.PLoS Genet 11, e1005091. |
| 170 | Wang XY, Wang XL, Hu QN, Dai XM, Tian HN, Zheng KJ, Wang XP, Mao TL, Chen JG, Wang SC (2015q). Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Ara- bidopsis.Plant J 83, 300-311. |
| 171 | Wang Y, Li K, Chen L, Zou Y, Liu H, Tian Y, Li D, Wang R, Zhao F, Ferguson BJ, Gresshoff PM, Li X (2015r). MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development.Plant Physiol 168, 984-999. |
| 172 | Wang Y, Li Y, Xue H, Pritchard HW, Wang X (2015s). Reactive oxygen species-provoked mitochondriadepen- dent cell death during ageing of elm (Ulmus pumila L.) seeds.Plant J 81, 438-452. |
| 173 | Wang YH, Jiang WM, Comes HP, Hu FS, Qiu YX, Fu CX (2015t). Molecular phylogeography and ecological niche modelling of a widespread herbaceous climber, Tetra- stigma hemsleyanum (Vitaceae): insights into Plio-Pleis- tocene range dynamics of evergreen forest in subtropical China.New Phytol 206, 852-867. |
| 174 | Wang YQ, Ma H (2015). Step-wise and lineage-specific diversification of plant RNA polymerase genes and origin of the largest plant-specific subunits.New Phytol 207, 1198-1212. |
| 175 | Wang YX, Xiong GS, Hu J, Jiang L, Yu H, Xu J, Fang YX, Zeng LJ, Xu EB, Xu J, Ye WJ, Meng XB, Liu RF, Chen HQ, Jing YH, Wang YH, Zhu XD, Li JY, Qian Q (2015u). Copy number variation at the GL7 locus contributes to grain size diversity in rice.Nat Genet 47, 944-948. |
| 176 | Wang Z, Ji H, Yuan B, Wang S, Su C, Yao B, Zhao H, Li X (2015v). ABA signaling is fine-tuned by antagonistic HAB1 variants.Nat Commun 6, 8138. |
| 177 | Wang Z, Zhou ZK, Liu YF, Liu TF, Li Q, Ji YY, Li CC, Fang C, Wang M, Wu M, Shen YT, Tang T, Ma JX, Tian ZX (2015w). Functional evolution of phosphatidylethanol-am- ine binding proteins in soybean and Arabidopsis.Plant Cell 27, 323-336. |
| 178 | Wang ZJ, Li HJ, Han ZF, Zhang HQ, Wang T, Lin GZ, Chang JB, Yang WC, Chai JJ (2015x). Allosteric recep- tor activation by the plant peptide hormone phytosul- fokine.Nature 525, 265-268. |
| 179 | Wei X, Liu KY, Zhang YX, Feng Q, Wang LH, Zhao Y, Li DH, Zhao Q, Zhu XD, Zhu XF, Li WJ, Fan DL, Gao Y, Lu YQ, Zhang XM, Tang XM, Zhou CC, Zhu CR, Liu LF, Zhong RC, Tian QL, Wen ZR, Weng QJ, Han B, Huang XH, Zhang XR (2015a). Genetic discovery for oil pro- duction and quality in sesame.Nat Commun 6, 8609. |
| 180 | Wei XP, Guo JT, Li M, Liu ZF (2015b). Structural mech- anism underlying the specific recognition between the Ar- abidopsis state-transition phosphatase TAP38/PPH1 and phosphorylated light-harvesting complex protein LHCB1.Plant Cell 27, 1113-1127. |
| 181 | Wen WW, Li K, Alseekh S, Omranian N, Zhao LJ, Zhou Y, Xiao YJ, Jin M, Yang N, Liu HJ, Florian A, Li WQ, Pan QC, Nikoloski Z, Yan JB, Fernie AR (2015). Genetic determinants of the network of primary metabolism and their relationships to plant performance in a maize re- combinant inbred line population.Plant Cell 27, 1839-1856. |
| 182 | Weng L, Zhao F, Li R, Xu C, Chen K, Xiao H (2015). The zinc finger transcription factor SlZFP2 negatively regul- ates abscisic acid biosynthesis and fruit ripening in toma- to.Plant Physiol 167, 931-949. |
| 183 | Wu G, Liu S, Zhao Y, Wang W, Kong Z, Tang D (2015a). ENHANCED DISEASE RESISTANCE4 associates with CLATHRIN HEAVY CHAIN2 and modulates plant imm- unity by regulating relocation of EDR1 in Arabidopsis.Plant Cell 27, 857-873. |
| 184 | Wu J, Nyman T, Wang DC, Argus GW, Yang YP, Chen JH (2015b). Phylogeny of Salix subgenus Salix s.l. (Salic- aceae): delimitation, biogeography, and reticulate evolu- tion.BMC Evol Biol 15, 31. |
| 185 | Wu J, Shahid MQ, Chen L, Chen Z, Wang L, Liu X, Lu Y (2015c). Polyploidy enhances F1 pollen sterility loci inter- actions that increase meiosis abnormalities and pollen sterility in autotetraploid rice.Plant Physiol 169, 2700-2717. |
| 186 | Wu J, Yang Z, Wang Y, Zheng L, Ye R, Ji Y, Zhao S, Ji S, Liu R, Xu L, Zheng H, Zhou Y, Zhang X, Cao X, Xie L, Wu Z, Qi Y, Li Y (2015d). Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by seq- uestering a host microRNA.Elife 4, e05733. |
| 187 | Wu JX, Li J, Liu Z, Yin J, Chang ZY, Rong C, Wu JL, Bi FC, Yao N (2015e). The Arabidopsis ceramidase AtACER functions in disease resistance and salt tolerance.Plant J 81, 767-780. |
| 188 | Wu SY, Xie YR, Zhang JJ, Ren YL, Zhang X, Wang JL, Guo XP, Wu FQ, Sheng PK, Wang J, Wu CY, Wang HY, Huang SJ, Wan JM (2015f). VLN2 regulates plant arch- itecture by affecting microfilament dynamics and polar auxin transport in rice.Plant Cell 27, 2829-2845. |
| 189 | Xia J, Niu S, Ciais P, Janssens IA, Chen J, Ammann C, Arain A, Blanken PD, Cescatti A, Bonal D, Buchmann N, Curtis PS, Chen S, Dong J, Flanagan LB, Frankenberg C, Georgiadis T, Gough CM, Hui D, Kiely G, Li J, Lund M, Magliulo V, Marcolla B, Merbold L, Montagnani L, Moors EJ, Olesen JE, Piao S, Raschi A, Roupsard O, Suyker AE, Urbaniak M, Vaccari FP, Varlagin A, Vesala T, Wilkinson M, Weng E, Wohlfahrt G, Yan L, Luo Y (2015). Joint control of terrestrial gross primary productivity by plant phenology and physiology.Proc Natl Acad Sci USA 112, 2788-2793. |
| 190 | Xiang QP, Wei R, Shao YZ, Yang ZY, Wang XQ, Zhang XC (2015). Phylogenetic relationships, possible ancient hyb- ridization, and biogeographic history of Abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes.Mol Phylogenet Evol 82PtA, 1-14. |
| 191 | Xiao L, Yang G, Zhang L, Yang X, Zhao S, Ji Z, Zhou Q, Hu M, Wang Y, Chen M, Xu Y, Jin H, Xiao X, Hu G, Bao F, Hu Y, Wan P, Li L, Deng X, Kuang T, Xiang C, Zhu JK, Oliver MJ, He Y (2015). The resurrection genome of Boea hygrometrica: a blueprint for survival of dehydration.Proc Natl Acad Sci USA 112, 5833-5837. |
| 192 | Xie WB, Wang GW, Yuan M, Yao W, Lyu K, Zhao H, Yang M, Li PB, Zhang X, Yuan J, Wang QX, Liu F, Dong HX, Zhang LJ, Li XL, Meng XZ, Zhang W, Xiong LZ, He YQ, Wang SP, Yu SB, Xu CG, Luo J, Li XH, Xiao JH, Lian XM, Zhang QF (2015). Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection.Proc Natl Acad Sci USA 112, E5411-E5419. |
| 193 | Xing J, Wang T, Liu Z, Xu J, Yao Y, Hu Z, Peng H, Xin M, Yu F, Zhou D, Ni Z (2015). GENERAL CONTROL NON- REPRESSED PROTEIN5-mediated histone acetylation of FERRIC REDUCTASE DEFECTIVE3 contributes to iron homeostasis in Arabidopsis.Plant Physiol 168, 1309-1320. |
| 194 | Xu C, Zhou X, Wen CK (2015a). HYPER RECOMBINA- TION1 of the THO/TREX complex plays a role in con- trolling transcription of the REVERSION-TO-ETHYLENE SENSITIVITY1 gene in Arabidopsis.PLoS Genet 11, e1004956. |
| 195 | Xu H, Detto M, Fang S, Li Y, Zang R, Liu S (2015b). Habitat hotspots of common and rare tropical species along climatic and edaphic gradients.J Ecol 103, 1325-1333. |
| 196 | Xu Z, Ren H, Li MH, van Ruijven J, Han X, Wan S, Li H, Yu Q, Jiang Y, Jiang L (2015c). Environmental changes drive the temporal stability of semi-arid natural grasslands through altering species asynchrony.J Ecol 103, 1308-1316. |
| 197 | Yan D, Zhang X, Zhang L, Ye S, Zeng L, Liu J, Li Q, He Z (2015a). CURVED CHIMERIC PALEA 1 encoding an EMF1-like protein maintains epigenetic repression of Os- MADS58 in rice palea development.Plant J 82, 12-24. |
| 198 | Yan S, McLamore ES, Dong S, Gao H, Taguchi M, Wang N, Zhang T, Su X, Shen Y (2015b). The role of plasma membrane H(+) -ATPase in jasmonate-induced ion fluxes and stomatal closure in Arabidopsis thaliana.Plant J 83, 638-649. |
| 199 | Yang H, Chang F, You C, Cui J, Zhu G, Wang L, Zheng Y, Qi J, Ma H (2015a). Whole-genome DNA methylation patterns and complex associations with gene structure and expression during flower development in Arabidopsis.Plant J 81, 268-281. |
| 200 | Yang L, Li B, Zheng XY, Li J, Yang M, Dong X, He G, An C, Deng XW (2015b). Salicylic acid biosynthesis is enhanced and contributes to increased biotrophic patho- gen resistance in Arabidopsis hybrids.Nat Commun 6, 7309. |
| 201 | Yang SG, Li CL, Zhao LM, Gao SJ, Lu JX, Zhao ML, Chen CY, Liu XC, Luo M, Cui YH, Yang CW, Wu KQ (2015c). The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance.Plant Cell 27, 1670-1680. |
| 202 | Yang SH, Wang L, Huang J, Zhang XH, Yuan Y, Chen JQ, Hurst LD, Tian DC (2015d). Parent-progeny sequencing indicates higher mutation rates in heterozygotes.Nature 523, 463-467. |
| 203 | Yang X, Huang Z, Zhang K, Cornelissen JHC (2015e). C:N:P stoichiometry of artemisia species and close rela- tives across northern China: unravelling effects of climate, soil and taxonomy.J Ecol 103, 1020-1031. |
| 204 | Yang X, Zhao XG, Li CQ, Liu J, Qiu ZJ, Dong Y, Wang YZ (2015f). Distinct regulatory changes underlying differential expression of TEOSINTE BRANCHED1-CYCLOIDEA- PROLIFERATING CELL FACTOR genes associated with petal variations in zygomorphic flowers of Petrocosmea spp. of the family Gesneriaceae.Plant Physiol 169, 2138-2151. |
| 205 | Yang Y, Fu D, Zhu C, He Y, Zhang H, Liu T, Li X, Wu C (2015g). The RING-finger ubiquitin ligase HAF1 mediates Heading date 1 degradation during photoperiodic flow- ering in rice.Plant Cell 27, 2455-2468. |
| 206 | Ye CY, Chen L, Liu C, Zhu QH, Fan L (2015). Widespread noncoding circular RNAs in plants.New Phytol 208, 88-95. |
| 207 | Yi TS, Jin GH, Wen J (2015). Chloroplast capture and intra- and inter-continental biogeographic diversification in the Asian-New World disjunct plant genus Osmorhiza (Apia- ceae).Mol Phylogenet Evol 85, 10-21. |
| 208 | Yin CC, Ma B, Collinge DP, Pogson BJ, He SJ, Xiong Q, Duan KX, Chen H, Yang C, Lu X, Wang YQ, Zhang WK, Chu CC, Sun XH, Fang S, Chu JF, Lu TG, Chen SY, Zhang JS (2015). Ethylene responses in rice roots and coleoptiles are differentially regulated by a carotenoid isomerase-mediated abscisic acid pathway.Plant Cell 27, 1061-1081. |
| 209 | Yu Q, Wilcox K, La Pierre K, Knapp AK, Han X, Smith MD (2015). Stoichiometric homeostasis predicts plant species dominance, temporal stability, and responses to global change.Ecology 96, 2328-2335. |
| 210 | Zeng XQ, Long H, Wang Z, Zhao SC, Tang YW, Huang ZY, Wang YL, Xu QJ, Mao LK, Deng GB, Yao XM, Li XF, Bai LJ, Yuan HJ, Pan ZF, Liu RJ, Chen X, WangMu QM, Chen M, Yu LL, Liang JJ, DunZhu DW, Zheng Y, Yu SY, Luobu ZX, Guang XM, Li J, Deng C, Hu WS, Chen CH, TaBa XN, Gao LY, Lü XD, Ben Abu Y, Fang XD, Nevo E, Yu MQ, Wang J, Tashi N (2015a). The draft genome of Tibetan hulless barley reveals adaptive patter- ns to the high stressful Tibetan Plateau.Proc Natl Acad Sci USA 112, 1095-1100. |
| 211 | Zeng YF, Wang WT, Liao WJ, Wang HF, Zhang DY (2015b). Multiple glacial refugia for cool-temperate deci- duous trees in northern East Asia: the Mongolian oak as a case study.Mol Ecol 24, 5676-5691. |
| 212 | Zeng YL, Chung KP, Li BY, Lai CM, Lam SK, Wang XF, Cui Y, Gao CJ, Luo M, Wong KB, Schekman R, Jiang LW (2015c). Unique COPII component AtSar1a/AtSec23a pair is required for the distinct function of protein ER ex- port in Arabidopsis thaliana.Proc Natl Acad Sci USA 112, 14360-14365. |
| 213 | Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q, Li C (2015). Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis.Plant Cell 27, 2814-2828. |
| 214 | Zhan X, Qian B, Cao F, Wu W, Yang L, Guan Q, Gu X, Wang P, Okusolubo TA, Dunn SL, Zhu JK, Zhu J (2015). An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA res- ponses.Nat Commun 6, 8139. |
| 215 | Zhang B, Liu C, Wang YQ, Yao X, Wang F, Wu JS, King GJ, Liu KD (2015a). Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower col- our from white to yellow in Brassica species.New Phytol 206, 1513-1526. |
| 216 | Zhang C, Cao L, Rong L, An Z, Zhou W, Ma J, Shen WH, Zhu Y, Dong A (2015b). The chromatin-remodeling factor AtINO80 plays crucial roles in genome stability main- tenance and in plant development.Plant J 82, 655-668. |
| 217 | Zhang CX, Chen CH, Dong HX, Shen JR, Dau H, Zhao JQ (2015c). A synthetic Mn4Ca-cluster mimicking the oxy- gen-evolving center of photosynthesis.Science 348, 690-693. |
| 218 | Zhang HW, Cui F, Wu YR, Lou LJ, Liu L, Tian MM, Ning Y, Shu K, Tang SY, Xie Q (2015d). The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis.Plant Cell 27, 214-227. |
| 219 | Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang HB (2015e). The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESIST- ANT3 and modulates iron homeostasis in Arabidopsis.Plant Cell 27, 787-805. |
| 220 | Zhang J, Zhou X, Yan W, Zhang Z, Lu L, Han Z, Zhao H, Liu H, Song P, Hu Y, Shen G, He Q, Guo S, Gao G, Wang G, Xing Y (2015f). Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice.New Phytol 208, 1056-1066. |
| 221 | Zhang TQ, Lian H, Tang H, Dolezal K, Zhou CM, Yu S, Chen JH, Chen Q, Liu H, Ljung K, Wang JW (2015g). An intrinsic microRNA timer regulates progressive decline in shoot regenerative capacity in plants.Plant Cell 27, 349-360. |
| 222 | Zhang X, Sun J, Cao X, Song XW (2015h). Epigenetic mutation of RAV6 affects leaf angle and seed size in rice.Plant Physiol 169, 2118-2128. |
| 223 | Zhang X, Yang S, Wang J, Jia Y, Huang J, Tan S, Zhong Y, Wang L, Gu L, Chen JQ, Pan Q, Bergelson J, Tian D (2015i). A genome-wide survey reveals abundant rice blast R genes in resistant cultivars.Plant J 84, 20-28. |
| 224 | Zhang XY, Zhu Y, Liu XD, Hong XY, Xu Y, Zhu P, Shen Y, Wu HH, Ji YS, Wen X, Zhang C, Zhao Q, Wang YC, Lu J, Guo HW (2015j). Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Ara- bidopsis.Science 348, 120-123. |
| 225 | Zhang Y, Du L, Xu R, Cui R, Hao J, Sun C, Li Y (2015k). Transcription factors SOD7/NGAL2 and DPA4/NGAL3 act redundantly to regulate seed size by directly repressing KLU expression in Arabidopsis thaliana.Plant Cell 27, 620-632. |
| 226 | Zhang Y, Jiao Y, Liu Z, Zhu YX (2015l). ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells.Nat Commun 6, 6003. |
| 227 | Zhang Y, Tan L, Zhu Z, Yuan L, Xie D, Sun C (2015m). TOND1 confers tolerance to nitrogen deficiency in rice.Plant J 81, 367-376. |
| 228 | Zhang Z, Yang J, Wu Y (2015n). Transcriptional regulation of Zein gene expression in maize through the additive and synergistic action of opaque2, prolamine-box binding fac- tor, and O2 heterodimerizing proteins.Plant Cell 27, 1162-1172. |
| 229 | Zhang ZH, Mao LY, Chen HM, Bu FJ, Li GC, Sun JJ, Li S, Sun HH, Jiao C, Blakely R, Pan JS, Cai R, Luo RB, Van de Peer Y, Jacobsen E, Fei ZJ, Huang SW (2015o). Genome-wide mapping of structural variations reveals a copy number variant that determines reproductive mor- phology in cucumber.Plant Cell 27, 1595-1604. |
| 230 | Zhao J, Chen HY, Ren D, Tang HW, Qiu R, Feng JL, Long YM, Niu BX, Chen DP, Zhong TY, Liu YG, Guo JX (2015a). Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Ory- za sativa).New Phytol 208, 936-948. |
| 231 | Zhao L, Tan L, Zhu Z, Xiao L, Xie D, Sun C (2015b). PAY1 improves plant architecture and enhances grain yield in rice.Plant J 83, 528-536. |
| 232 | Zhao M, Yang S, Chen CY, Li C, Shan W, Lu W, Cui Y, Liu X, Wu K (2015c). Arabidopsis BREVIPEDICELLUS inter- acts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture.PLoS Genet 11, e1005125. |
| 233 | Zhao S, Wu Y, He Y, Wang Y, Xiao J, Li L, Wang Y, Chen X, Xiong W, Wu Y (2015d). RopGEF2 is involved in ABA-suppression of seed germination and post-germina- tion growth of Arabidopsis.Plant J 84, 886-899. |
| 234 | Zhao Y, Cheng S, Song Y, Huang Y, Zhou S, Liu X, Zhou DX (2015e). The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling.Plant Cell 27, 2469-2483. |
| 235 | Zhao Y, Hu Y, Dai M, Huang L, Zhou DX (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice.Plant Cell 21, 736-748. |
| 236 | Zheng M, Wang Y, Wang Y, Wang C, Ren Y, Lü J, Peng C, Wu T, Liu K, Zhao S, Liu X, Guo X, Jiang L, Terzaghi W, Wan J (2015). DEFORMED FLORAL ORGAN1 (DFO1) regulates floral organ identity by epigenetically repressing the expression of OsMADS58 in rice (Oryza sativa).New Phytol 206, 1476-1490. |
| 237 | Zhong C, Xu H, Ye S, Wang S, Li L, Zhang S, Wang X (2015). Gibberellic acid-stimulated Arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis.Plant Physiol 169, 2288-2303. |
| 238 | Zhou H, Kui LW, Wang HL, Gu C, Dare AP, Espley RV, He HP, Allan AC, Han YP (2015a). Molecular genetics of blood-fleshed peach reveals activation of anthocyanin bio- synthesis by NAC transcription factors.Plant J 82, 105-121. |
| 239 | Zhou ML, Sun ZM, Wang CL, Zhang XQ, Tang YX, Zhu XM, Shao JR, Wu YM (2015b). Changing a conserved amino acid in R2R3-MYB transcription repressors results in cytoplasmic accumulation and abolishes their repres- sive activity in Arabidopsis.Plant J 84, 395-403. |
| 240 | Zhou SF, Sun L, Valdes AE, Engstrom P, Song ZT, Lu SJ, Liu JX (2015c). Membrane-associated transcription factor peptidase, site-2 protease, antagonizes ABA signaling in Arabidopsis.New Phytol 208, 188-197. |
| 241 | Zhou W, Barrett SCH, Wang H, Li DZ (2015d). Reciprocal herkogamy promotes disassortative mating in a distylous species with intramorph compatibility.New Phytol 206, 1503-1512. |
| 242 | Zhou Z, Shi H, Chen B, Zhang R, Huang S, Fu Y (2015e). Arabidopsis RIC1 severs actin filaments at the apex to regulate pollen tube growth.Plant Cell 27, 1140-1161. |
| 243 | Zhou Z, Wu Y, Yang Y, Du M, Zhang X, Guo Y, Li C, Zhou JM (2015f). An Arabidopsis plasma membrane proton ATPase modulates JA signaling and is exploited by the pseudomonas syringae effector protein AvrB for stomatal invasion.Plant Cell 27, 2032-2041. |
| 244 | Zhu H, Fu B, Wang S, Zhu L, Zhang L, Jiao L, Wang C (2015a). Reducing soil erosion by improving community functional diversity in semi-arid grasslands.J Appl Ecol 52, 1063-1072. |
| 245 | Zhu XY, Chen JY, Xie ZK, Gao J, Ren GD, Gao S, Zhou X, Kuai BK (2015b). Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regula- tion of major chlorophyll catabolic genes.Plant J 84, 597-610. |
| 246 | Zhu Y, Li Y, Fei F, Wang Z, Wang W, Cao A, Liu Y, Han S, Xing L, Wang H, Chen W, Tang S, Huang X, Shen Q, Xie Q, Wang X (2015c). E3 ubiquitin ligase gene CMPG1-V from Haynaldia villosa L. contributes to pow- dery mildew resistance in common wheat (Triticum aes- tivum L.).Plant J 84, 154-168. |
| 247 | Zou LH, Huang JX, Zhang GQ, Liu ZJ, Zhuang XY (2015). A molecular phylogeny of Aeridinae (Orchidaceae: Ep- idendroideae) inferred from multiple nuclear and chloro- plast regions.Mol Phylogenet Evol 85, 247-254. |
| 248 | Zuo W, Chao Q, Zhang N, Ye J, Tan G, Li B, Xing Y, Zhang B, Liu H, Fengler KA, Zhao J, Zhao X, Chen Y, Lai J, Yan J, Xu M (2015). A maize wall-associated kinase confers quantitative resistance to head smut.Nat Genet 47, 151-157. |
| [1] | Peng-Bin Han Kang-Di PEI Bin He Shu-Li XIAO Cindy Q.TANG. Community composition and characteristics of Tsuga dumosa forests in China [J]. Chin J Plant Ecol, 2025, 49(植被): 1-0. |
| [2] | Bin He Peng-Bin Han Shu-Li XIAO Kang-Di PEI Cindy Q.TANG. Community types and characteristics of Keteleeria davidiana forests [J]. Chin J Plant Ecol, 2025, 49(植被): 1-0. |
| [3] | Menglu Wei, Jinyue Li, Yuchun Rao. Practice of Cultivating Innovative Talents in the Field of Plant Science Based on "Promoting Innovation through Competition" [J]. Chinese Bulletin of Botany, 2025, 60(4): 1-0. |
| [4] | Tian Zhiqi, Su Yang. China’s implementation model of international environmental conventions and its application to the Convention on Biological Diversity—Achieving the Kunming-Montreal Global Biodiversity Framework targets and the role of national parks [J]. Biodiv Sci, 2025, 33(3): 24593-. |
| [5] | Zhou Zhihua, Jin Xiaohua, Luo Ying, Li Diqiang, Yue Jianbing, Liu Fang, He Tuo, Li Xi, Dong Hui, Luo Peng. Analyses and suggestions on mechanisms of forestry and grassland administrations in China to achieve targets of Kunming-Montreal Global Biodiversity Framework [J]. Biodiv Sci, 2025, 33(3): 24487-. |
| [6] | Hongya Gu, Fan Chen, Rongcheng Lin, Xiaoquan Qi, Shuhua Yang, Zhiduan Chen, Xuewei Chen, Zhaojun Ding, Langtao Xiao, Jianru Zuo, Liwen Jiang, Yongfei Bai, Kang Chong, Lei Wang. Achievements and Advances of Plant Sciences Research in China in 2024 [J]. Chinese Bulletin of Botany, 2025, 60(2): 151-171. |
| [7] | Wei Yi, Yi Ai, Meng Wu, Liming Tian, Tserang Donko Mipam. Soil archaeal community responses to different grazing intensities in the alpine meadows of the Qinghai-Tibetan Plateau [J]. Biodiv Sci, 2025, 33(1): 24339-. |
| [8] | Bangze Li, Shuren Zhang. An updated species checklist and taxonomic synopsis of Cyperaceae in China [J]. Biodiv Sci, 2024, 32(7): 24106-. |
| [9] | Zonggang Hu. China and the United States once planned joint compilation of the Flora of China after the victory in the China’s Resistance War Against Japan [J]. Biodiv Sci, 2024, 32(6): 24220-. |
| [10] | Qiyu Kuang, Liang Hu. Species diversity and geographical distribution of marine benthic shell-bearing mollusks around Donghai Island and Naozhou Island, Guangdong Province [J]. Biodiv Sci, 2024, 32(5): 24065-. |
| [11] | Yujie Chi, Xintian Zhang, Zhixuan Tian, Chengshuai Guan, Xinzhi Gu, Zhihui Liu, Zhanbin Wang, Jinjie Wang. Species diversity of powdery mildew fungi (Erysiphaceae) and their host plants in Northeast Asia [J]. Biodiv Sci, 2024, 32(4): 23443-. |
| [12] | PAN Yuan-Fang, PAN Liang-Hao, QIU Si-Ting, QIU Guang-Long, SU Zhi-Nan, SHI Xiao-Fang, FAN Hang-Qing. Variations in tree height among mangroves and their environmental adaptive mechanisms in China’s coastal areas [J]. Chin J Plant Ecol, 2024, 48(4): 483-495. |
| [13] | Fan Chen, Hongya Gu, Xiaoquan Qi, Rongcheng Lin, Qian Qian, Langtao Xiao, Shuhua Yang, Jianru Zuo, Yongfei Bai, Zhiduan Chen, Zhaojun Ding, Xiaojing Wang, Liwen Jiang, Kang Chong, Lei Wang. Achievements and Advances of Plant Sciences Research in China in 2023 [J]. Chinese Bulletin of Botany, 2024, 59(2): 171-187. |
| [14] | Di Lin, Shuanglin Chen, Que Du, Wenlong Song, Gu Rao, Shuzhen Yan. Investigation of species diversity of myxomycetes in Dabie Mountains [J]. Biodiv Sci, 2024, 32(2): 23242-. |
| [15] | Huanxi Cao, Qingsong Zhou, Arong Luo, Pu Tang, Tingjing Li, Zejian Li, Huayan Chen, Zeqing Niu, Chaodong Zhu. New taxa of extant Hymenoptera in 2023 [J]. Biodiv Sci, 2024, 32(11): 24319-. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||