

Chinese Bulletin of Botany ›› 2016, Vol. 51 ›› Issue (3): 387-395.DOI: 10.11983/CBB15113 cstr: 32102.14.CBB15113
• SPECIAL TOPICS • Previous Articles Next Articles
Bodan Su, Jinxing Lin, Jianwei Xiao*
Received:2015-06-18
Accepted:2015-11-01
Online:2016-05-01
Published:2016-05-24
Contact:
Xiao Jianwei
About author:? These authors contributed equally to this paper
Bodan Su, Jinxing Lin, Jianwei Xiao. Ubiquitin-mediated Membrane Protein Trafficking in Plant Cells and Related Research Techniques: A Review[J]. Chinese Bulletin of Botany, 2016, 51(3): 387-395.
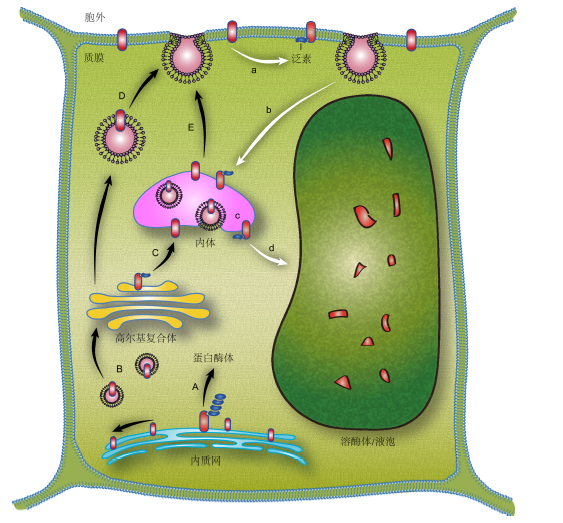
Fig. 1 Cellular ubiquitin-mediated membrane protein trafficking processes((a)-(d) Ub-dependent internalization/sorting of membrane protein; (A)-(E) Ub-dependent internalization/sorting of newly synthesized proteins)
| 检测方法 | 体内检测 | 体外检测 | ||
|---|---|---|---|---|
| 生物化学法 | 细胞生物学法 | 分子与生物化学法 | ||
| 主要步骤 | 特异性抗体免疫沉淀并富集靶蛋白; 特异性抗体检测蛋白泛素化以及类型; 质谱分析技术检测蛋白泛素化位点 | 蛋白链接荧光标签转入植株; 利用显微镜辅以蛋白泛素化抑制剂处理检测蛋白前后变化; 单分子水平研究蛋白分子的运动与转运 | 利用酵母双杂交筛库技术筛出特异性E3蛋白; 外源表达E1、E2、E3和目标蛋白并孵育; 特异性抗体检测蛋白泛素化以及类型、质谱分析技术检测蛋白泛素化位点 | |
| 优点 | 特异性强, 周期短, 操作简单 | 活体观察, 简单、直观, 可量化信息 | 可大量表达蛋白, 周期短 | |
| 缺点 | 膜蛋白易降解, 样品纯度不高 | 周期较长, 用量较多, 需要稳定且有功能的材料 | 有“假阴性”结果, 无法精确模拟体内环境 | |
Table 1 Comparison of different methods for detecting the membrane proteins ubiquitination
| 检测方法 | 体内检测 | 体外检测 | ||
|---|---|---|---|---|
| 生物化学法 | 细胞生物学法 | 分子与生物化学法 | ||
| 主要步骤 | 特异性抗体免疫沉淀并富集靶蛋白; 特异性抗体检测蛋白泛素化以及类型; 质谱分析技术检测蛋白泛素化位点 | 蛋白链接荧光标签转入植株; 利用显微镜辅以蛋白泛素化抑制剂处理检测蛋白前后变化; 单分子水平研究蛋白分子的运动与转运 | 利用酵母双杂交筛库技术筛出特异性E3蛋白; 外源表达E1、E2、E3和目标蛋白并孵育; 特异性抗体检测蛋白泛素化以及类型、质谱分析技术检测蛋白泛素化位点 | |
| 优点 | 特异性强, 周期短, 操作简单 | 活体观察, 简单、直观, 可量化信息 | 可大量表达蛋白, 周期短 | |
| 缺点 | 膜蛋白易降解, 样品纯度不高 | 周期较长, 用量较多, 需要稳定且有功能的材料 | 有“假阴性”结果, 无法精确模拟体内环境 | |
| [1] | Amerik AY, Nowak J, Swaminathan S, Hochstrasser M (2000). The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic path- ways.Mol Biol Cell 11, 3365-3380. |
| [2] | Apaja PM, Xu H, Lukacs GL (2010). Quality control for unfolded proteins at the plasma membrane.J Cell Biol 191, 553-570. |
| [3] | Avila-Ospina L, Moison M, Yoshimoto K, Masclaux- Daubresse C (2014). Autophagy, plant senescence, and nutrient recycling.J Exp Bot 65, 3799-3811. |
| [4] | Bagola K, Mehnert M, Jarosch E, Sommer T (2011). Protein dislocation from the ER.BBA-Biomembranes 1808, 925-936. |
| [5] | Bar M, Schuster S, Leibman M, Ezer R, Avni A (2014). The function of EHD2 in endocytosis and defense signaling is affected by SUMO.Plant Mol Biol 84, 509-518. |
| [6] | Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants.Proc Natl Acad Sci USA 108, E450-E458. |
| [7] | Battey NH, James NC, Greenland AJ, Brownlee C (1999). Exocytosis and endocytosis.Plant Cell 11, 643-659. |
| [8] | Bilodeau PS, Winistorfer SC, Allaman MM, Surendhran K, Kearney WR, Robertson AD, Piper RC (2004). The GAT domains of clathrin-associated GGA proteins have two ubiquitin binding motifs.J Biol Chem 279, 54808-54816. |
| [9] | Bonnemaison ML, Eipper BA, Mains RE (2013). Role of adaptor proteins in secretory granule biogenesis and maturation.Front Neuroendocrinol 4, 101. |
| [10] | Brown NG, VanderLinden R, Watson ER, Qiao R, Grace CR, Yamaguchi M, Weissmann F, Frye JJ, Dube P, Cho SE (2015). RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex.Proc Natl Acad Sci USA 112, 5272-5279. |
| [11] | Cowles CR, Odorizzi G, Payne GS, Emr SD (1997). The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole.Cell 91, 109-118. |
| [12] | Crapeau M, Merhi A, André B (2014). Stress conditions promote yeast Gap1 permease ubiquitylation and down- regulation via the arrestin-like bul and aly proteins.J Biol Chem 289, 22103-22116. |
| [13] | Cui F, Liu L, Zhao Q, Zhang Z, Li Q, Lin B, Wu Y, Tang S, Xie Q (2012). Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid- mediated salt stress tolerance. Plant Cell 24, 233-244. |
| [14] | Demmel L, Gravert M, Ercan E, Habermann B, Müller- Reichert T, Kukhtina V, Haucke V, Baust T, Sohrmann M, Kalaidzidis Y (2008). The clathrin adaptor Gga2p is a phosphatidylinositol 4-phosphate effector at the Golgi exit.Mol Biol Cell 19, 1991-2002. |
| [15] | Denic V, Quan EM, Weissman JS (2006). A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation.Cell 126, 349-359. |
| [16] | Donoso G, Herzog V, Schmitz A (2005). Misfolded BiP is degraded by a proteasorne-independent endoplasmic- reticulum-associated degradation pathway.B Journal 387, 897-903. |
| [17] | Dores MR, Chen B, Lin H, Soh UJ, Paing MM, Montagne WA, Meerloo T, Trejo J (2012). ALIX binds a YPX3L motif of the GPCR PAR1 and mediates ubiquitin-ind- ependent ESCRT-III/MVB sorting.J Cell Biol 197, 407-419. |
| [18] | Ellgaard L, Helenius A (2003). Quality control in the endoplasmic reticulum.Nat Rev Mol Cell Biol 4, 181-191. |
| [19] | Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weiss- man AM (2001). The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum.Proc Natl Acad Sci USA 98, 14422-14427. |
| [20] | Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB.Curr Biol 18, 1824-1832. |
| [21] | Goldstein G, Scheid M, Hammerling U, Schlesinger D, Niall H, Boyse E (1975). Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells.Proc Natl Acad Sci USA 72, 11-15. |
| [22] | Guo Y, Sirkis DW, Schekman R (2014). Protein sorting at the trans-Golgi Network.Annu Rev Cell Dev Biol 30, 169-206. |
| [23] | Haglund K, Dikic I (2012). The role of ubiquitylation in receptor endocytosis and endosomal sorting.J Cell Sci 125, 265-275. |
| [24] | Han L, Yang J, Wang X, Wu Q, Yin S, Li Z, Zhang J, Xing Y, Chen Z, Tsun A (2014). The E3 deubiquitinase USP17 is a positive regulator of retinoic acid-related orphan nuclear receptor γt (RORγt) in Th17 Cells.J Biol Chem 289, 25546-25555. |
| [25] | Hicke L, Dunn R (2003). Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19, 141-172. |
| [26] | Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T (2009). The ubiquitylation machinery of the endoplasmic reticulum.Nature 458, 453-460. |
| [27] | Hong E, Davidson AR, Kaiser CA (1996). A pathway for targeting soluble misfolded proteins to the yeast vacuole.J Cell Biol 135, 623-633. |
| [28] | Hurley JH, Stenmark H (2011). Molecular mechanisms of ubiquitin-dependent membrane traffic.Annu Rev Biophys 40, 119. |
| [29] | Ikeda F, Dikic I (2008). A typical ubiquitin chains: new molecular signals.EMBO Rep 9, 536-542. |
| [30] | Johansen T, Lamark T (2011). Selective autophagy mediated by autophagic adapter proteins.Autophagy 7, 279-296. |
| [31] | Kölling R, Losko S (1997). The linker region of the ABC- transporter Ste6 mediates ubiquitination and fast turnover of the protein.EMBO J 16, 2251-2261. |
| [32] | Keener JM, Babst M (2013). Quality control and substrate-dependent downregulation of the nutrient transporter Fur4.Traffic 14, 412-427. |
| [33] | Kirkham M, Parton RG (2005). Clathrin-independent endocytosis: new insights into caveolae and non-caveolar lipid raft carriers.Biochim Biophys Acta 1745, 273-286. |
| [34] | Kirkin V, McEwan DG, Novak I, Dikic I (2009). A role for ubiquitin in selective autophagy.Mol Cell 34, 259-269. |
| [35] | Lauwers E, Jacob C, André B (2009). K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway.J Cell Biol 185, 493-502. |
| [36] | Lemus L, Goder V (2014). Regulation of endoplasmic reticulum-associated protein degradation (ERAD) by ubiquitin.Cells 3, 824-847. |
| [37] | Lin WY, Huang TK, Chiou TJ (2013). NITROGEN LIMITATION ADAPTATION, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Ara- bidopsis.Plant Cell 25, 4061-4074. |
| [38] | Liu L, Cui F, Li Q, Yin B, Zhang H, Lin B, Wu Y, Xia R, Tang S, Xie Q (2011). The endoplasmic reticulum- associated degradation is necessary for plant salt tolerance.Cell Res 21, 957-969. |
| [39] | Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, Zhang H, Dong L, Guo H, Xie Q (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana.Plant J 61, 893-903. |
| [40] | Liu Y, Bassham DC (2012). Autophagy: pathways for self-eating in plant cells.Annu Rev Plant Biol 63, 215-237. |
| [41] | Luhtala N, Odorizzi G (2004). Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes.J Cell Biol 166, 717-729. |
| [42] | MacGurn JA, Hsu PC, Emr SD (2012). Ubiquitin and membrane protein turnover: from cradle to grave.Annu Rev Biochem 81, 231-259. |
| [43] | Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS (2013). The ubiquitin ligase parkin mediates resistance to intracellular pathogens.Nature 501, 512-516. |
| [44] | Marchese A (2015). Monitoring chemokine receptor trafficking by confocal immunofluorescence microscopy.Method Enzymol 570, 281-292. |
| [45] | Martins S, Dohmann EM, Cayrel A, Johnson A, Fischer W, Pojer F, Satiat-Jeunemaître B, Jaillais Y, Chory J, Geldner N (2015). Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination.Nat Commun 6, 6151. |
| [46] | Mergner J, Heinzlmeir S, Kuster B, Schwechheimer C (2015). DENEDDYLASE1 deconjugates NEDD8 from non- cullin protein substrates in Arabidopsis thaliana.Plant Cell 27, 741-753. |
| [47] | Mishra SK, Ammon T, Popowicz GM, Krajewski M, Nagel RJ, Ares M, Holak TA, Jentsch S (2011). Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.Nature 474, 173-178. |
| [48] | Mukhopadhyay D, Riezman H (2007). Proteasome-indep endent functions of ubiquitin in endocytosis and signaling.Science 315, 201-205. |
| [49] | Parton RG, Richards AA (2003). Lipid rafts and caveolae as portals for endocytosis: new insights and common mechanisms.Traffic 4, 724-738. |
| [50] | Passmore L, Barford D (2004). Getting into position: the catalytic mechanisms of protein ubiquitylation.Biochem J 379, 513-525. |
| [51] | Petroski MD, Salvesen GS, Wolf DA (2011). Urm1 couples sulfur transfer to ubiquitin-like protein function in oxidative stress.Proc Natl Acad Sci USA 108, 1749-1750. |
| [52] | Pickart CM, Eddins MJ (2004). Ubiquitin: structures, functions, mechanisms.BBA-Mol Cell Res 1695, 55-72. |
| [53] | Piper RC, Dikic I, Lukacs GL (2014). Ubiquitin-dependent sorting in endocytosis.Cold Spring Harb Perspect Biol 6, a016808. |
| [54] | Puertollano R, Bonifacino JS (2004). Interactions of GGA3 with the ubiquitin sorting machinery.Nat Cell Biol 6, 244-251. |
| [55] | Ravid T, Hochstrasser M (2008). Diversity of degradation signals in the ubiquitin-proteasome system.Nat Rev Mol Cell Biol 9, 679-689. |
| [56] | Scheuring D, Künzl F, Viotti C, Yan MSW, Jiang L, Sch- ellmann S, Robinson DG, Pimpl P (2012). Ubiquitin initiates sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway.BMC Plant Biol 12, 164. |
| [57] | Scott D, Oldham NJ, Strachan J, Searle MS, Layfield R (2014). Ubiquitin-binding domains: mechanisms of ubiquitin recognition and use as tools to investigate ubiquitin-modified proteomes.Proteomics 15, 844-861. |
| [58] | Seaman MN (2012). The retromer complex-endosomal pro- tein recycling and beyond.J Cell Sci 125, 4693-4702. |
| [59] | Shields SB, Piper RC (2011). How ubiquitin functions with ESCRTs.Traffic 12, 1306-1317. |
| [60] | Shih SC, Sloper-Mould KE, Hicke L (2000). Monoubiquitin carries a novel internalization signal that is appended to activated receptors.EMBO J 19, 187-198. |
| [61] | Shin LJ, Lo JC, Chen GH, Callis J, Fu H, Yeh KC (2013). IRT1 degradation factor1, a ring E3 ubiquitin ligase, regu- lates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 25, 3039-3051. |
| [62] | Starr TL, Pagant S, Wang CW, Schekman R (2012). Sorting signals that mediate traffic of chitin synthase III bet- ween the TGN/endosomes and to the plasma membrane in yeast.PLoS One 7, e46386. |
| [63] | Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415-3428. |
| [64] | Su W, Liu Y, Xia Y, Hong Z, Li J (2011). Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis.Proc Natl Acad Sci USA 108, 870-875. |
| [65] | Terrell J, Shih S, Dunn R, Hicke L (1998). A function for monoubiquitination in the internalization of a G protein-coupled receptor.Mol Cell 1, 193-202. |
| [66] | van der Veen AG, Ploegh HL (2012). Ubiquitin-like proteins.Annu Rev Biochem 81, 323-357. |
| [67] | Vembar SS, Brodsky JL (2008). One step at a time: endoplasmic reticulum-associated degradation.Nat Rev Mol Cell Biol 9, 944-957. |
| [68] | Wang S, Thibault G, Ng DT (2011). Routing misfolded proteins through the multivesicular body (MVB) pathway protects against proteotoxicity.J Biol Chem 286, 29376-29387. |
| [69] | Wenzel D, Stoll K, Klevit R (2011). E2s: structurally economical and functionally replete.Biochem J 433, 31-42. |
| [70] | Weyand S, Shimamura T, Yajima S, Suzuki Si, Mirza O, Krusong K, Carpenter EP, Rutherford NG, Hadden JM, O'Reilly J (2008). Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter.Science 322, 709-713. |
| [71] | Wyatt D, Malik R, Vesecky AC, Marchese A (2011). Small ubiquitin-like modifier modification of arrestin-3 regulates receptor trafficking.J Biol Chem 286, 3884-3893. |
| [72] | Zhang X, Garreton V, Chua NH (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation.Genes Dev 19, 1532-1543. |
| [73] | Zhao Q, Tian M, Li Q, Cui F, Liu L, Yin B, Xie Q (2013). A plant-specific in vitro ubiquitination analysis system.Plant J 74, 524-533. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||