基因编辑技术在玉米中的研究进展
- 山东农业大学, 泰安 271018
†共同第一作者
收稿日期: 2024-05-27
录用日期: 2024-08-20
网络出版日期: 2024-08-28
基金资助
山东省自然科学基金(ZR202211230214)
Research Progress of Gene Editing Technology in Maize
- Shandong Agricultural University, Tai’an 271018, China
†These authors contributed equally to this paper
Received date: 2024-05-27
Accepted date: 2024-08-20
Online published: 2024-08-28
摘要
基因编辑技术已成为现代农业育种领域的重要工具。玉米(Zea mays)是全球最重要的粮食作物之一, 基因编辑技术在玉米中的应用显示出巨大的潜力。该文综述了基因编辑技术在玉米研究中的应用进展, 重点介绍了CRISPR/Cas等系统在玉米基因组编辑中的最新成果。首先, 介绍了基因编辑技术的基本原理和类型, 特别是CRISPR/Cas系统的工作机制及其在玉米中的应用优势。其次, 总结了基因编辑技术在玉米育种中的研究进展, 涵盖从基础的基因组编辑到复杂的多基因编辑, 旨在改良玉米的产量、品质和抗逆性等关键性状。最后, 文章列举了我国在玉米基因编辑方面的杰出工作, 并讨论了基因编辑技术应用于玉米育种中存在的问题, 同时展望了未来发展方向。
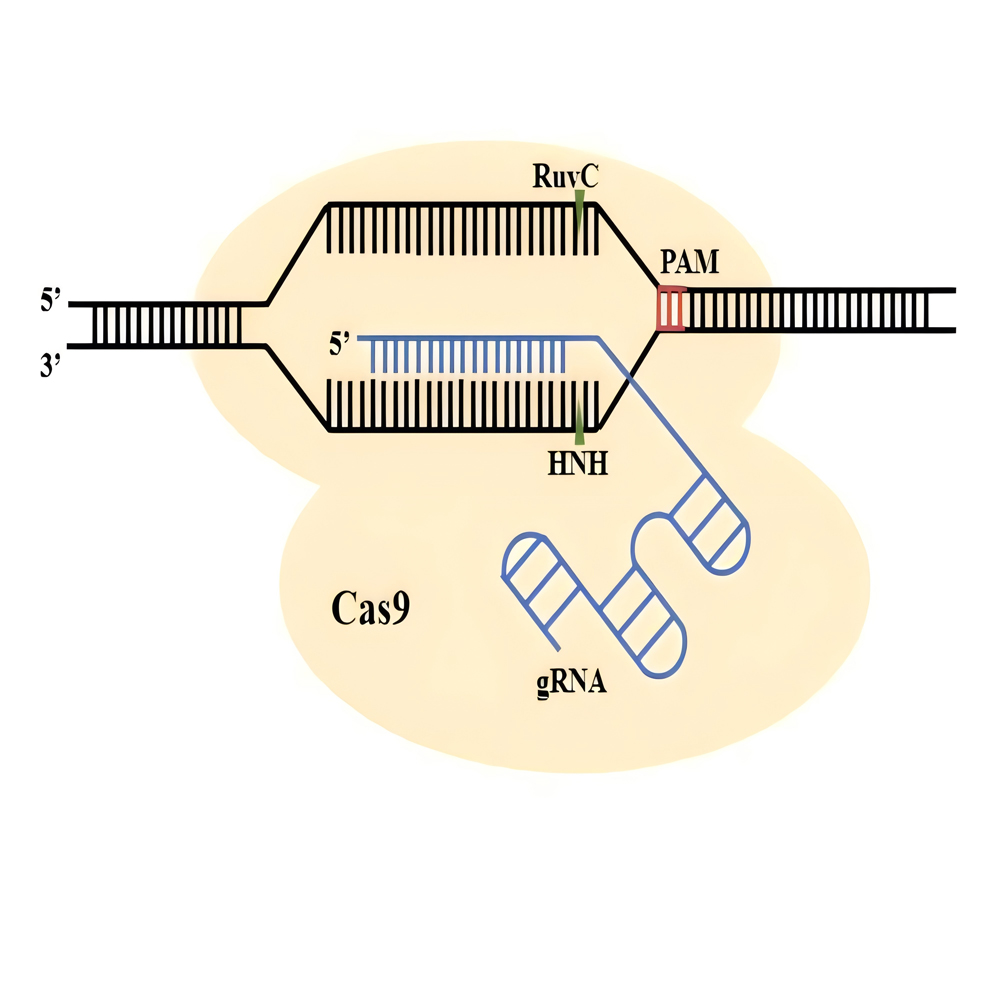
关键词: 玉米; 基因组编辑; CRISPR/Cas系统; 遗传改良
本文引用格式
张强 , 赵振宇 , 李平华 . 基因编辑技术在玉米中的研究进展[J]. 植物学报, 2024 , 59(6) : 978 -998 . DOI: 10.11983/CBB24080
Abstract
Gene editing technology has become an important tool in crop breeding. Maize, one of the globally most important food crops, has been shown with great potential in the use of gene editing technology in genome research and breeding. In this paper, we reviewed the recent progress and applications of gene editing technology in maize research, with a focus on the latest achievements in maize genome editing by CRISPR/Cas. Firstly, we introduced the basic principles and types of gene editing technology, particularly the working mechanism of the CRISPR/Cas systems, and its application advantages in maize. Secondly, we summarized the research progress of gene editing technology in maize breeding, from basic genome editing to the editing of complex multi-gene regulation, aiming at the improvement of key traits such as yield, grain quality, and stress resistance. Finally, the outstanding research work in maize gene editing in China is presented and the existing issues of gene editing technology in maize breeding are discussed, along with an outlook on future development trends.

Key words: maize; genome editing; CRISPR/Cas system; genetic improvement
参考文献
| [1] | Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding SW, Mahfouz M (2018). RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol 19, 1. |
| [2] | Anand A, Bass SH, Wu E, Wang N, Mcbride KE, Annaluru N, Miller M, Hua M, Jones TJ (2018). An improved ternary vector system for Agrobacterium-mediated rapid maize transformation. Plant Mol Biol 97, 187-200. |
| [3] | Bai MY, Lin WX, Peng CY, Song PZ, Kuang HQ, Lin JN, Zhang JP, Wang JY, Chen B, Li HR, Kong FJ, Jia GF, Guan YF (2024a). Expressing a human RNA demethylase as an assister improves gene-editing efficiency in plants. Mol Plant 17, 363-366. |
| [4] | Bai YH, Liu SN, Bai Y, Xu ZS, Zhao HN, Zhao HM, Lai JS, Liu Y, Song WB (2024b). Application of CRISPR/Cas12i.3 for targeted mutagenesis in broomcorn millet (Panicum miliaceum L.). J Integr Plant Biol 66, 1544-1547. |
| [5] | Bandyopadhyay A, Kancharla N, Javalkote VS, Dasgupta S, Brutnell TP (2020). CRISPR-Cas12a (Cpf1): a versatile tool in the plant genome editing tool box for agricultural advancement. Front Plant Sci 11, 584151. |
| [6] | Bezrutczyk M, Hartwig T, Horschman M, Char SN, Yang JL, Yang B, Frommer WB, Sosso D (2018). Impaired phloem loading in zmsweet13a, b, c sucrose transporter triple knock-out mutants in Zea mays. New Phytol 218, 594-603. |
| [7] | Butiuc-Keul A, Farkas A, Carpa R, Iordache D (2022). CRISPR-Cas system: the powerful modulator of accessory genomes in prokaryotes. Microb Physiol 32, 2-17. |
| [8] | Cao XS, Xie HT, Song ML, Zhao LH, Deng S, Tian YF, Li GF, Lang ZB, Zhu JK (2023). Extremely simplified cut- dip-budding method for genetic transformation and gene editing in Taraxacum kok-saghyz. Innovation Life 1, 100040. |
| [9] | Cathomen T, Joung JK (2008). Zinc-finger nucleases: the next generation emerges. Mol Ther 16, 1200-1207. |
| [10] | Chen F, Gu HY, Qi XQ, Lin RC, Qian Q, Xiao LT, Yang SH, Zuo JR, Bai YF, Chen ZD, Ding ZJ, Wang XJ, Jiang LW, Chong K, Wang L (2024). Achievements and advances of plant sciences research in China in 2023. Chin Bull Bot 59, 171-187. (in Chinese) |
| 陈凡, 顾红雅, 漆小泉, 林荣呈, 钱前, 萧浪涛, 杨淑华, 左建儒, 白永飞, 陈之端, 丁兆军, 王小菁, 姜里文, 种康, 王雷 (2024). 2023年中国植物科学重要研究进展. 植物学报 59, 171-187. | |
| [11] | Chen L, Zhang S, Xue NN, Hong MJ, Zhang XH, Zhang D, Yang J, Bai SJ, Huang YF, Meng HW, Wu H, Luan CM, Zhu BY, Ru GM, Gao HY, Zhong LP, Liu MZ, Liu MY, Cheng YY, Yi CQ, Wang LR, Zhao YX, Song GJ, Li DL (2023). Engineering a precise adenine base editor with minimal bystander editing. Nat Chem Biol 19, 101-110. |
| [12] | Chen PJ, Hussmann JA, Yan J, Knipping F, Ravisankar P, Chen PF, Chen CD, Nelson JW, Newby GA, Sahin M, Osborn MJ, Weissman JS, Adamson B, Liu DR (2021). Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635-5652. |
| [13] | Chen RR, Xu QL, Liu Y, Zhang JJ, Ren DT, Wang GY, Liu YJ (2018). Generation of transgene-free maize male sterile lines using the CRISPR/Cas9 system. Front Plant Sci 9, 1180. |
| [14] | Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823. |
| [15] | Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F (2017). RNA editing with CRISPR-Cas13. Science 358, 1019-1027. |
| [16] | Doman JL, Pandey S, Neugebauer ME, An MR, Davis JR, Randolph PB, Mcelroy A, Gao XD, Raguram A, Richter MF, Everette KA, Banskota S, Tian K, Tao YA, Tolar J, Osborn MJ, Liu DR (2023). Phage-assisted evolution and protein engineering yield compact, efficient prime editors. Cell 186, 3983-4002. |
| [17] | Doman JL, Raguram A, Newby GA, Liu DR (2020). Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat Biotechnol 38, 620-628. |
| [18] | Dong L, Li LN, Liu CL, Liu CX, Geng SF, Li XH, Huang CL, Mao L, Chen SJ, Xie CX (2018). Genome editing and double-fluorescence proteins enable robust maternal haploid induction and identification in maize. Mol Plant 11, 1214-1217. |
| [19] | Dong L, Qi XT, Zhu JJ, Liu CL, Zhang X, Cheng BJ, Mao L, Xie CX (2019). Supersweet and waxy: meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol J 17, 1853-1855. |
| [20] | Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gooten- berg JS, Yozwiak NL, Zhang F, Sabeti PC (2019). Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell 76, 826-837. |
| [21] | Gao HR, Gadlage MJ, Lafitte HR, Lenderts B, Yang MZ, Schroder M, Farrell J, Snopek K, Peterson D, Feigenbutz L, Jones S, St Clair G, Rahe M, Sanyour- Doyel N, Peng CN, Wang LJ, Young JK, Beatty M, Dahlke B, Hazebroek J, Greene TW, Cigan AM, Chilcoat ND, Meeley RB (2020). Superior field performance of waxy corn engineered using CRISPR-Cas9. Nat Biotechnol 38, 579-581. |
| [22] | Gaudelli NM, Lam DK, Rees HA, Solá-Esteves NM, Barrera LA, Born DA, Edwards A, Gehrke JM, Lee SJ, Liquori AJ, Murray R, Packer MS, Rinaldi C, Slaymaker IM, Yen J, Young LE, Ciaramella G (2020). Directed evolution of adenine base editors with increased activity and therapeutic application. Nat Biotechnol 38, 892-900. |
| [23] | Gong P, Bontinck M, Demuynck K, De Block J, Gevaert K, Eeckhout D, Persiau G, Aesaert S, Coussens G, Van Lijsebettens M, Pauwels L, De Jaeger G, Inzé D, Nelissen H (2022). SAMBA controls cell division rate during maize development. Plant Physiol 188, 411-424. |
| [24] | Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F (2017). Nucleic acid detection with CRISPR- Cas13a/C2c2. Science 356, 438-442. |
| [25] | Grünewald J, Zhou RH, Iyer S, Lareau CA, Garcia SP, Aryee MJ, Joung JK (2019). CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat Biotechnol 37, 1041-1048. |
| [26] | Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J (2018). CRISPR-Cas9 genome editing induces a p53- mediated DNA damage response. Nat Med 24, 927-930. |
| [27] | He XL, Liu PC, Ma BJ, Chen XF (2022). Advance in gene- editing technology based on CRISPR/Cas9 and its application in plants. Chin Bull Bot 57, 508-531. (in Chinese) |
| 何晓玲, 刘鹏程, 马伯军, 陈析丰 (2022). 基于CRISPR/ Cas9的基因编辑技术研究进展及其在植物中的应用. 植物学报 57, 508-531. | |
| [28] | Hille F, Richter H, Wong SP, Bratovi? M, Ressel S, Charpentier E (2018). The biology of CRISPR-Cas: backward and forward. Cell 172, 1239-1259. |
| [29] | Hu JC, Sun Y, Li BS, Liu Z, Wang ZW, Gao Q, Guo MY, Liu GW, Zhao KT, Gao CX (2024). Strand-preferred base editing of organellar and nuclear genomes using CyDENT. Nat Biotechnol 42, 936-945. |
| [30] | Huang J, Lu G, Liu L, Raihan MS, Xu JT, Jian LM, Zhao LX, Tran TM, Zhang QH, Liu J, Li WQ, Wei CX, Braun DM, Li Q, Fernie AR, Jackson D, Yan JB (2020). The Kernel size-related quantitative trait locus qKW9 encodes a pentatricopeptide repeat protein that affects photosynthesis and grain filling. Plant Physiol 183, 1696-1709. |
| [31] | Huang JY, Lin QP, Fei HY, He ZX, Xu H, Li YJ, Qu KL, Han P, Gao Q, Li BS, Liu GW, Zhang LX, Hu JC, Zhang R, Zuo EW, Luo YL, Ran YD, Qiu JL, Zhao KT, Gao CX (2023). Discovery of deaminase functions by structure- based protein clustering. Cell 186, 3182-3195. |
| [32] | Jiang YL, Li ZW, Liu XZ, Zhu TT, Xie K, Hou QC, Yan TW, Niu CF, Zhang SW, Yang MB, Xie RR, Wang J, Li JP, An XL, Wan XY (2021). ZmFAR1 and ZmABCG26 regulated by microRNA are essential for lipid metabolism in maize anther. Int J Mol Sci 22, 7916. |
| [33] | Jiang YY, Chai YP, Lu MH, Han XL, Lin QP, Zhang Y, Zhang Q, Zhou Y, Wang XC, Gao CX, Chen QJ (2020). Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol 21, 257. |
| [34] | Jin S, Zong Y, Gao Q, Zhu ZX, Wang YP, Qin P, Liang CZ, Wang DW, Qiu JL, Zhang F, Gao CX (2019). Cytosine, but not adenine, base editors induce genome-wide off- target mutations in rice. Science 364, 292-295. |
| [35] | Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821. |
| [36] | Jinek M, Jiang FG, Taylor DW, Sternberg SH, Kaya E, Ma EB, Anders C, Hauer M, Zhou KH, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA (2014). Structures of Cas9 endonucleases reveal RNA- mediated conformational activation. Science 343, e1247997. |
| [37] | Kaul T, Thangaraj A, Jain R, Bharti J, Kaul R, Verma R, Sony SK, Abdel Motelb KF, Yadav P, Agrawal PK (2024). CRISPR/Cas9-mediated homology donor repair base editing system to confer herbicide resistance in maize (Zea mays L.). Plant Physiol Biochem 207, 108374. |
| [38] | Kelliher T, Starr D, Su XJ, Tang GZ, Chen ZY, Carter J, Wittich PE, Dong SJ, Green J, Burch E, Mccuiston J, Gu WN, Sun YJ, Strebe T, Roberts J, Bate NJ, Que QD (2019). One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol 37, 287-292. |
| [39] | Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR (2017). Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 35, 371-376. |
| [40] | Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, Maianti JP, Raguram A, Liu DR (2018). Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol 36, 843-846. |
| [41] | Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420-424. |
| [42] | Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, Kim YB, Badran AH, Liu DR (2017). Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 3, eaao4774. |
| [43] | Kurt IC, Zhou RH, Iyer S, Garcia SP, Miller BR, Langner LM, Grünewald J, Joung JK (2021). CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol 39, 41-46. |
| [44] | Lee HK, Smith HE, Liu CY, Willi M, Hennighausen L (2020). Cytosine base editor 4 but not adenine base editor generates off-target mutations in mouse embryos. Commun Biol 3, 19. |
| [45] | Li CX, Liu CL, Qi XT, Wu YC, Fei XH, Mao L, Cheng BJ, Li XH, Xie CX (2017a). RNA-guided Cas9 as an in vivo desired-target mutator in maize. Plant Biotechnol J 15, 1566-1576. |
| [46] | Li J, Zhang HW, Si XM, Tian YH, Chen KL, Liu JX, Chen HB, Gao CX (2017b). Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J Genet Genomics 44, 465-468. |
| [47] | Li JF, Norville JE, Aach J, Mccormack M, Zhang DD, Bush J, Church GM, Sheen J (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31, 688-691. |
| [48] | Li N, Sun Y, Yang LY (2013). Cytological study of pollen-mediated plant transformation method based on GFP observation. Chin Bull Bot 48, 616-622. (in Chinese) |
| 李娜, 孙毅, 杨利艳 (2013). 基于GFP观察的花粉介导植物转基因方法的细胞学研究. 植物学报 48, 616-622. | |
| [49] | Li QQ, Wu GX, Zhao YP, Wang BB, Zhao BB, Kong DX, Wei HB, Chen CX, Wang HY (2020). CRISPR/Cas9- mediated knockout and overexpression studies reveal a role of maize phytochrome C in regulating flowering time and plant height. Plant Biotechnol J 18, 2520-2532. |
| [50] | Li WT, He M, Chen XW (2019). Discovery of ZmFBL41Chang7-2 as a key weapon against banded leaf and sheath blight resistance in maize. Chin Bull Bot 54, 547-549. (in Chinese) |
| 李伟滔, 贺闽, 陈学伟 (2019). ZmFBL41Chang7-2: 玉米抗纹枯病的关键利器. 植物学报 54, 547-549. | |
| [51] | Li X (2018). Optimization of the Genetic Transformation System of Caladium bicolor ‘Hongtao K’ and the Introduction of AtPAP1 Gene and Maize Lc Gene. Master’s thesis. Chongqing: Southwest University. (in Chinese) |
| 李想 (2018). 彩叶芋‘红桃K’遗传转化体系的优化及AtPAP1基因与玉米Lc基因的导入. 硕士论文. 重庆: 西南大学. | |
| [52] | Liang RH, He ZX, Zhao KT, Zhu HC, Hu JC, Liu GW, Gao Q, Liu MY, Zhang R, Qiu JL, Gao CX (2024). Prime editing using CRISPR-Cas12a and circular RNAs in human cells. Nat Biotechnol doi: 10.1038/s41587-023-02095-x. |
| [53] | Liu C, Li Y, Dai PH, Yao ZP, Liu XD (2020). Construction of genome editing vectors targeting CKXs in maize. Mol Plant Breed 18, 7051-7055. (in Chinese) |
| 刘超, 李月, 代培红, 姚正培, 刘晓东 (2020). 玉米CKXs基因组编辑载体的构建. 分子植物育种 18, 7051-7055. | |
| [54] | Liu CL, Kong M, Yang F, Zhu JJ, Qi XT, Weng JF, Di DP, Xie CX (2022). Targeted generation of null mutants in ZmGDIα confers resistance against maize rough dwarf disease without agronomic penalty. Plant Biotechnol J 20, 803-805. |
| [55] | Liu CX, Li X, Meng DX, Zhong Y, Chen C, Dong X, Xu XW, Chen BJ, Li W, Li L, Tian XL, Zhao HM, Song WB, Luo HS, Zhang QH, Lai JS, Jin WW, Yan JB, Chen SJ (2017). A 4bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol Plant 10, 520-522. |
| [56] | Liu J, Yan JB (2019). A teosinte rare allele increases maize plant density and yield. Chin Bull Bot 54, 554-557. (in Chinese) |
| 刘杰, 严建兵 (2019). 大刍草稀有等位基因促进玉米密植高产. 植物学报 54, 554-557. | |
| [57] | Liu L, Gallagher J, Arevalo ED, Chen R, Skopelitis T, Wu QY, Bartlett M, Jackson D (2021). Enhancing grain- yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat Plants 7, 287-294. |
| [58] | Liu WY, Lin HH, Yu CP, Chang CK, Chen HJ, Lin JJ, Lu MYJ, Tu SL, Shiu SH, Wu SH, Ku MSB, Li WH (2020). Maize ANT1 modulates vascular development, chloroplast development, photosynthesis, and plant growth. Proc Natl Acad Sci USA 117, 21747-21756. |
| [59] | Liu XS, Gu DF, Zhang YR, Jiang YL, Xiao Z, Xu RF, Qin RY, Li J, Wei PC (2024). Conditional knockdown of OsMLH1 to improve plant prime editing systems without disturbing fertility in rice. Genome Biol 25, 131. |
| [60] | Lowe K, La Rota M, Hoerster G, Hastings C, Wang N, Chamberlin M, Wu E, Jones T, Gordon-Kamm W (2018). Rapid genotype "independent" Zea mays L. (maize) trans- formation via direct somatic embryogenesis. In Vitro Cell Dev Biol Plant 54, 240-252. |
| [61] | Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang LJ, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao ZM, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng PZ, Bidney D, Falco C, Register J, Zhao ZY, Xu DP, Jones T, Gordon-Kamm W (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998-2015. |
| [62] | Luo Y, Zhang ML, Liu Y, Liu J, Li WQ, Chen GS, Peng Y, Jin M, Wei WJ, Jian LM, Yan J, Fernie AR, Yan JB (2022). Genetic variation in YIGE1 contributes to ear length and grain yield in maize. New Phytol 234, 513-526. |
| [63] | Lv P, Su F, Chen FY, Yan CX, Xia DD, Sun H, Li SS, Duan ZQ, Ma CL, Zhang H, Wang MG, Niu XM, Zhu JK, Zhang JS (2024). Genome editing in rice using CRISPR/ Cas12i3. Plant Biotechnol J 22, 379-385. |
| [64] | Ma L, Sun YL, Ruan XS, Huang PC, Wang S, Li SF, Zhou Y, Wang F, Cao Y, Wang Q, Wang ZH, Kolomiets MV, Gao XQ (2021). Genome-wide characterization of jasmonates signaling components reveals the essential role of ZmCOI1a-ZmJAZ15 action module in regulating maize immunity to gibberella stalk rot. Int J Mol Sci 22, 870. |
| [65] | Mahmud S, Ahmed J, Aziz MA, Hasan MR, Shaon SM, Bhuiyan MNAH, Rahman MF, Rakib HH, Islam MS (2017). Clustered regularly interspaced short palindromic repeats Cas systems: a comprehensive review. Int J Basic Clin Pharmacol 4, 613-622. |
| [66] | Mali P, Yang LH, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM (2013). RNA-guided human genome engineering via Cas9. Science 339, 823-826. |
| [67] | Mishra R, Joshi RK, Zhao KJ (2020). Base editing in crops: current advances, limitations and future implications. Plant Biotechnol J 18, 20-31. |
| [68] | Mladenov E, Iliakis G (2011). Induction and repair of DNA double strand breaks: the increasing spectrum of non- homologous end joining pathways. Mutat Res 711, 61-72. |
| [69] | Mu LY (2022). Creation of Gene Editing Materials for Three Maize 100-Kernel Weight Candidate Genes. Master’s thesis. Wuhan: Huazhong Agricultural University. (in Chinese). |
| 穆路遥 (2022). 三个玉米百粒重候选基因基因编辑材料的创制. 硕士论文. 武汉: 华中农业大学. | |
| [70] | Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31, 691-693. |
| [71] | Nelson JW, Randolph PB, Shen SP, Everette KA, Chen PJ, Anzalone AV, An MR, Newby GA, Chen JC, Hsu A, Liu DR (2022). Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol 40, 402-410. |
| [72] | Ning Q, Jian YN, Du YF, Li YF, Shen XM, Jia HT, Zhao R, Zhan JM, Yang F, Jackson D, Liu L, Zhang ZX (2021). An ethylene biosynthesis enzyme controls quantitative variation in maize ear length and kernel yield. Nat Commun 12, 5832. |
| [73] | Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, Shimatani Z, Kondo A (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, 1248. |
| [74] | Pan ZY, Liu M, Zhao HL, Tan ZD, Liang K, Sun Q, Gong DM, He HJ, Zhou WQ, Qiu FZ (2020). ZmSRL5 is involved in drought tolerance by maintaining cuticular wax structure in maize. J Integr Plant Biol 62, 1895-1909. |
| [75] | Pathi KM, Rink P, Budhagatapalli N, Betz R, Saado I, Hiekel S, Becker M, Djamei A, Kumlehn J (2020). Engineering smut resistance in maize by site-directed mutagenesis of LIPOXYGENASE 3. Front Plant Sci 11, 543895. |
| [76] | Qi XT (2019). Engineering Liguleless, Super-Sweet and Waxy Compound Mazie Varieties via CRISPR-Cas9. PhD disseration. Hefei: Anhui Agricultural University. (in Chinese) |
| 祁显涛 (2019). 基于CRISPR-Cas9的玉米紧凑株型与甜糯复合型性状定向遗传改良. 博士论文. 合肥: 安徽农业大学. (in Chinese) | |
| [77] | Qi XT, Wu H, Jiang HY, Zhu JJ, Huang CL, Zhang X, Liu CL, Cheng BJ (2020). Conversion of a normal maize hybrid into a waxy version using in vivo CRISPR/Cas9 targeted mutation activity. Crop J 8, 440-448. |
| [78] | Qiao DX, Wang JY, Lu MH, Xin CP, Chai YP, Jiang YY, Sun W, Cao ZH, Guo SY, Wang XC, Chen QJ (2023). Optimized prime editing efficiently generates heritable mutations in maize. J Integr Plant Biol 65, 900-906. |
| [79] | Ren JJ, Wu PH, Trampe B, Tian XL, Lübberstedt T, Chen SJ (2017). Novel technologies in doubled haploid line development. Plant Biotechnol J 15, 1361-1370. |
| [80] | Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, Wilson C, Koblan LW, Zeng J, Bauer DE, Doudna JA, Liu DR (2020). Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol 38, 883-891. |
| [81] | Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635-644. |
| [82] | Shan QW, Wang YP, Li J, Zhang Y, Chen KL, Liang Z, Zhang K, Liu JX, Xi JJ, Qiu JL, Gao CX (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31, 686-688. |
| [83] | Shi JR, Gao HR, Wang HY, Lafitte HR, Archibald RL, Yang MZ, Hakimi SM, Mo H, Habben JE (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J 15, 207-216. |
| [84] | Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV (2015). Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell 60, 385-397. |
| [85] | Siebert R, Puchta H (2002). Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell 14, 1121-1131. |
| [86] | Sun C, Lei Y, Li BS, Gao Q, Li YJ, Cao W, Yang C, Li HC, Wang ZW, Li Y, Wang YP, Liu J, Zhao KT, Gao CX (2024). Precise integration of large DNA sequences in plant genomes using prime root editors. Nat Biotechnol 42, 316-327. |
| [87] | Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A (2016). Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 7, 13274. |
| [88] | Svitashev S, Young JK, Schwartz C, Gao HR, Falco SC, Cigan AM (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 169, 931-945. |
| [89] | Teng C, Zhang H, Hammond R, Huang K, Meyers BC, Walbot V (2020). Dicer-like 5 deficiency confers temperature-sensitive male sterility in maize. Nat Commun 11, 2912. |
| [90] | Wang BB, Zhu L, Zhao BB, Zhao YP, Xie YR, Zheng ZG, Li YY, Sun J, Wang HY (2019a). Development of a haploid-inducer mediated genome editing system for accelerating maize breeding. Mol Plant 12, 597-602. |
| [91] | Wang H, Yan SJ, Xin HJ, Huang WJ, Zhang H, Teng SZ, Yu YC, Fernie AR, Lu XD, Li PC, Li SY, Zhang CY, Ruan YL, Chen LQ, Lang ZH (2019b). A subsidiary cell-localized glucose transporter promotes stomatal conductance and photosynthesis. Plant Cell 31, 1328-1343. |
| [92] | Wang N, Arling M, Hoerster G, Ryan L, Wu E, Lowe K, Gordon-Kamm W, Jones TJ, Chilcoat ND, Anand A (2020). An efficient gene excision system in maize. Front Plant Sci 11, 1298. |
| [93] | Wang N, Gent JI, Dawe RK (2021a). Haploid induction by a maize cenh3 null mutant. Sci Adv 7, eabe2299. |
| [94] | Wang W, Pan QL, He F, Akhunova A, Chao S, Trick H, Akhunov E (2018). Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. Crispr J 1, 65-74. |
| [95] | Wang XD, Guo YH, Wang YR, Peng YL, Zhang HW, Zheng J (2024). ZmHDT103 negatively regulates drought stress tolerance in maize seedlings. Agronomy 14, 134. |
| [96] | Wang YX (2021). Using CRISPR/Cas Gene Editing Technology to Create Scented Corn. Master’s thesis. Jinan: Shandong Normal University. (in Chinese) |
| 王彦晓 (2021). 利用CRISPR/Cas基因编辑技术创建香味玉米. 硕士论文. 济南: 山东师范大学. (in Chinese) | |
| [97] | Wang YX, Liu XQ, Zheng XX, Wang WX, Yin XQ, Liu HF, Ma CL, Niu XM, Zhu JK, Wang F (2021b). Creation of aromatic maize by CRISPR/Cas. J Integr Plant Biol 63, 1664-1670. |
| [98] | Wei L, Zhang X, Zhang ZH, Liu HH, Lin ZW (2018). A new allele of the Brachytic2 gene in maize can efficiently modify plant architecture. Heredity 121, 75-86. |
| [99] | Wu GX, Zhao YP, Shen RX, Wang BB, Xie YR, Ma XJ, Zheng ZG, Wang HY (2019). Characterization of maize phytochrome-interacting factors in light signaling and photomorphogenesis. Plant Physiol 181, 789-803. |
| [100] | Xu JT, Liu XG, Jin ML, Pan H, Han BZ, Li MJ, Yan S, Hu GQ, Yan JB (2022). Establishment of genotype-independent high-efficiency transformation system in maize. Acta Agron Sin 48, 2987-2993. (in Chinese) |
| 许洁婷, 刘相国, 金敏亮, 潘弘, 韩宝柱, 李梦娇, 岩说, 胡国庆, 严建兵 (2022). 不依赖基因型的高效玉米遗传转化体系的建立. 作物学报 48, 2987-2993. | |
| [101] | Xu RB, Li YF, Sui Z, Lan TY, Song WJ, Zhang M, Zhang YR, Xing JW (2021). A C-terminal encoded peptide, ZmCEP1, is essential for kernel development in maize. J Exp Bot 72, 5390-5406. |
| [102] | Yan F, Kuang YJ, Ren B, Wang JW, Zhang DW, Lin HH, Yang B, Zhou XP, Zhou HB (2018). Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant 11, 631-634. |
| [103] | Yan J, Oyler-Castrillo P, Ravisankar P, Ward CC, Levesque S, Jing Y, Simpson D, Zhao AQ, Li H, Yan WH, Goudy L, Schmidt R, Solley SC, Gilbert LA, Chan MM, Bauer DE, Marson A, Parsons LR, Adamson B (2024). Improving prime editing with an endogenous small RNA- binding protein. Nature 628, 639-647. |
| [104] | Yang F, Li Y (2017). The new generation tool for CRISPR genome editing: CRISPR/Cpf1. Chin J Biotechnol 33, 361-371. (in Chinese) |
| 杨帆, 李寅 (2017). 新一代基因组编辑系统CRISPR/Cpf1. 生物工程学报 33, 361-371. | |
| [105] | Zhang JJ, Zhang XF, Chen RR, Yang L, Fan KJ, Liu Y, Wang GY, Ren ZJ, Liu YJ (2020a). Generation of transgene-free semidwarf maize plants by gene editing of Gibberellin-Oxidase20-3 using CRISPR/Cas9. Front Plant Sci 11, 1048. |
| [106] | Zhang X, Shi YX, Lu BS, Wu Y, Liu Y, Wang YD, Yang JX, Zhao JR (2021). Creation of new maize variety with fragrant rice like flavor by editing BADH2-1 and BADH2-2 using CRISPR/Cas9. Sci Agric Sin 54, 2064-2072. (in Chinese) |
| 张翔, 史亚兴, 卢柏山, 武莹, 刘亚, 王元东, 杨进孝, 赵久然 (2021). 利用CRISPR/Cas9技术编辑BADH2-1/BADH2-2创制香米味道玉米新种质. 中国农业科学 54, 2064-2072. | |
| [107] | Zhang Y, Zhang F, Li XH, Baller JA, Qi YP, Starker CG, Bogdanove AJ, Voytas DF (2013). Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 161, 20-27. |
| [108] | Zhang ZH, Zhang X, Lin ZL, Wang J, Liu HQ, Zhou LN, Zhong SY, Li Y, Zhu C, Lai JS, Li XR, Yu JM, Lin ZW (2020b). A large transposon insertion in the stiff1 promoter increases stalk strength in maize. Plant Cell 32, 152-165. |
| [109] | Zhou CY, Sun YD, Yan R, Liu YJ, Zuo EW, Gu C, Han LX, Wei Y, Hu XD, Zeng R, Li YX, Zhou HB, Guo F, Yang H (2019). Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571, 275-278. |
| [110] | Zong Y, Liu YJ, Xue CX, Li BS, Li XY, Wang YP, Li J, Liu GW, Huang XX, Cao XF, Gao CX (2022). An engineered prime editor with enhanced editing efficiency in plants. Nat Biotechnol 40, 1394-1402. |
| [111] | Zuo EW, Sun YD, Wei W, Yuan TL, Ying WQ, Sun H, Yuan LY, Steinmetz LM, Li YX, Yang H (2019). Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289-292. |