植物基因组学中的流式细胞分析及分选技术
- 华中农业大学生命科学技术学院, 武汉 430070
收稿日期: 2024-07-18
录用日期: 2024-08-20
网络出版日期: 2024-08-22
基金资助
国家重点研发计划(2023ZD04073);华中农业大学研究生培养条件建设(2024SC03)
Flow Cytometric Analysis and Sorting in Plant Genomics
- College of Life Science and Technology, Huazhong Agricultural University, Wuhan 430070, China
Received date: 2024-07-18
Accepted date: 2024-08-20
Online published: 2024-08-22
摘要
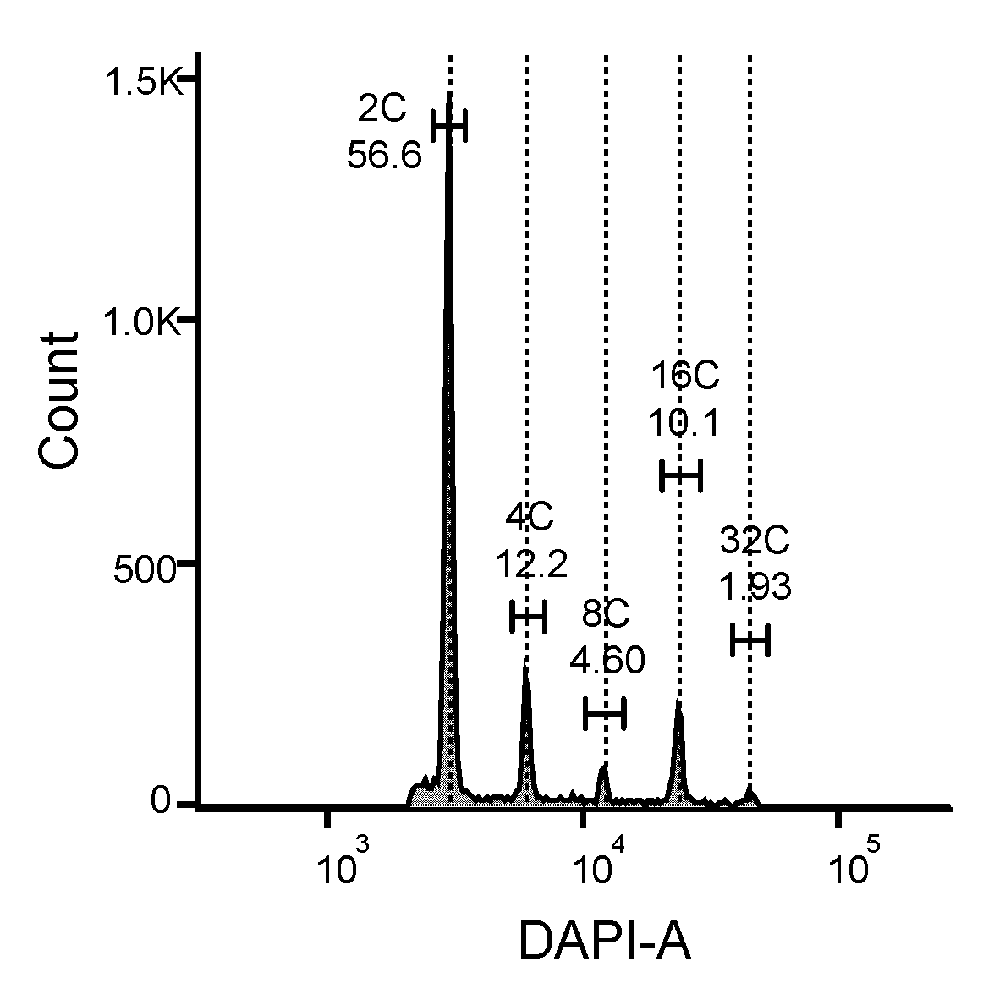
流式细胞术是一种高通量技术, 可以同时快速检测单个颗粒物的多项物理及生物学特征。随着测序成本的大幅降低, 流式细胞术在植物基因组学高通量样品获取中的作用日益凸显。该文以水稻(Oryza sativa)和大豆(Glycine max)为例, 详细介绍了应用流式细胞术对植物细胞核进行精细分选以及后续的ATAC-seq和RNA-seq实验分析流程, 为农业生物育种中基因的高效挖掘提供了优选工具。同时针对实验操作中的关键技术和常见问题, 如细胞核制备注意事项、分选纯度与效率的平衡及单细胞分选调试方法进行分析并提出建议, 为植物科学工作者应用流式细胞术开展基因组学研究提供参考。
本文引用格式
夏春皎 , 李运广 , 夏姝 , 庞伟 , 陈春丽 . 植物基因组学中的流式细胞分析及分选技术[J]. 植物学报, 2024 , 59(5) : 774 -782 . DOI: 10.11983/CBB24107
Abstract
Flow cytometry is a high-throughput technology that allows for the simultaneous and rapid detection of multiple physical and biological characteristics of individual particles. With the significant reduction in sequencing costs, flow cytometry is playing an increasingly prominent role in high-throughput sample acquisition for plant genomics. Taking rice and soybean as examples, this paper describes in detail the application of flow cytometry for fine sorting of plant cell nuclei and the subsequent ATAC-seq and RNA-seq experiments and analysis process, which provides a preferred tool for efficient mining of genes for agrobiological breeding. The key techniques and common problems in the experimental operation, such as the precautions for cell nuclei preparation, the balance between sorting purity and efficiency, and the debugging method for single-cell sorting, were also analyzed and suggested to provide references for the plant scientists in applying flow cytometry to carry out genomics research.

参考文献
| [1] | Bressan D, Battistoni G, Hannon GJ (2023). The dawn of spatial omics. Science 381, eabq4964. |
| [2] | Büscher M (2019). Flow cytometry instrumentation—an overview. Curr Protoc Cytom doi: 10.1002/cpcy.52. |
| [3] | Cápal P, Said M, Molnár I, Doležel J (2023). Flow cytometric analysis and sorting of plant chromosomes. In: Heitkam T, Garcia S, eds. Plant Cytogenetics and Cytogenomics. New York: Humana. pp. 177-200. |
| [4] | Decaestecker W, Bollier N, Buono RA, Nowack MK, Jacobs TB (2022). Protoplast preparation and fluorescence- activated cell sorting for the evaluation of targeted mutagenesis in plant cells. In: Wang K, Zhang F, eds. Protoplast Technology. New York: Humana. pp. 205-221. |
| [5] | Doležel J, Číhalíková J, Lucretti S (1992). A high-yield procedure for isolation of metaphase chromosomes from root tips of Vicia faba L. Planta 188, 93-98. |
| [6] | Doležel J, Göhde W (1995). Sex determination in dioecious plants Melandrium album and M. rubrum using high-resolution flow cytometry. Cytometry 19, 103-106. |
| [7] | Doležel J, Greilhuber J, Suda J (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2, 2233-2244. |
| [8] | Doležel J, Lucretti S, Molnár I, Cápal P, Giorgi D (2021). Chromosome analysis and sorting. Cytometry A 99, 328- 342. |
| [9] | Dpooležel J, Binarová P, Lcretti S (1989). Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plant 31, 113-120. |
| [10] | Fan W, Xia CJ, Wang SX, Liu J, Deng LJ, Sun SY, Wang XL (2022). Rhizobial infection of 4C cells triggers their endoreduplication during symbiotic nodule development in soybean. New Phytol 234, 1018-1030. |
| [11] | Galbraith DW, Afonso CL, Harkins KR (1984). Flow sorting and culture of protoplasts: conditions for high-frequency recovery, growth and morphogenesis from sorted protoplasts of suspension cultures of nicotiana. Plant Cell Rep 3, 151-155. |
| [12] | Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049-1051. |
| [13] | Grindberg RV, Yee-Greenbaum JL, McConnell MJ, Novotny M, O'Shaughnessy AL, Lambert GM, Araúzo- Bravo MJ, Lee J, Fishman M, Robbins GE, Lin XY, Venepally P, Badger JH, Galbraith DW, Gage FH, Lasken RS (2013). RNA-sequencing from single nuclei. Proc Natl Acad Sci USA 110, 19802-19807. |
| [14] | Guedes JG, Guimaraes AL, Carqueijeiro I, Gardner R, Bispo C, Sottomayor M (2022). Isolation of specialized plant cells by fluorescence-activated cell sorting. In: Fett-Neto AG,ed. Plant Secondary Metabolism Engineering. New York: Humana. pp. 193-200. |
| [15] | Guillotin B, Rahni R, Passalacqua M, Mohammed MA, Xu XS, Raju SK, Ramírez CO, Jackson D, Groen SC, Gillis J, Birnbaum KD (2023). A pan-grass transcriptome reveals patterns of cellular divergence in crops. Nature 617, 785-791. |
| [16] | Hang HY, Liu CC, Ren DD (2019). Development, application and prospection of flow cytometry. China Biotechnol 39(9), 68-83. (in Chinese) |
| 杭海英, 刘春春, 任丹丹 (2019). 流式细胞术的发展、应用及前景. 中国生物工程杂志 39(9), 68-83. | |
| [17] | Kamentsky LA, Melamed MR, Derman H (1965). Spectrophotometer: new instrument for ultrarapid cell analysis. Science 150, 630-631. |
| [18] | Kawakatsu T, Stuart T, Valdes M, Breakfield N, Schmitz RJ, Nery JR, Urich MA, Han XW, Lister R, Benfey PN, Ecker JR (2016). Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat Plants 2, 16058. |
| [19] | Li F, Hu Y, Wang F, Zhang Z, Liu XL, Bai SL, He YK (2010). Flow cytometry sorting of early developmental non- hair cells in roots of Arabidopsis thaliana. Chin Bull Bot 45, 460-465. (in Chinese) |
| 李斐, 胡勇, 王帆, 张珍, 刘祥林, 白素兰, 何奕昆 (2010). 利用流式细胞仪分选拟南芥根尖发育早期非根毛细胞. 植物学报 45, 460-465. | |
| [20] | Lin CS, Hsu CT, Yang LH, Lee LY, Fu JY, Cheng QW, Wu FH, Hsiao HCW, Zhang YS, Zhang R, Chang WJ, Yu CT, Wang W, Liao LJ, Gelvin SB, Shih MC (2018). Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J 16, 1295-1310. |
| [21] | Loureiro J, Rodriguez E, Doležel J, Santos C (2007). Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann Bot 100, 875-888. |
| [22] | Marx V (2016). Plants: a tool box of cell-based assays. Nat Methods 13, 551-554. |
| [23] | Mayer KFX, Martis M, Hedley PE, Šimková H, Liu H, Morris JA, Steuernagel B, Taudien S, Roessner S, Gundlach H, Kubaláková M, Suchánková P, Murat F, Felder M, Nussbaumer T, Graner A, Salse J, Endo T, Sakai H, Tanaka T, Itoh T, Sato K, Platzer M, Matsumoto T, Scholz U, Doležel J, Waugh R, Stein N (2011). Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23, 1249-1263. |
| [24] | Nitta N, Sugimura T, Isozaki A, Mikami H, Hiraki K, Sakuma S, Iino T, Arai F, Endo T, Fujiwaki Y, Fukuzawa H, Hase M, Hayakawa T, Hiramatsu K, Hoshino Y, Inaba M, Ito T, Karakawa H, Kasai Y, Koizumi K, Lee S, Lei C, Li M, Maeno T, Matsusaka S, Murakami D, Nakagawa A, Oguchi Y, Oikawa M, Ota T, Shiba K, Shintaku H, Shirasaki Y, Suga K, Suzuki Y, Suzuki N, Tanaka Y, Tezuka H, Toyokawa C, Yalikun Y, Yamada M, Yamagishi M, Yamano T, Yasumoto A, Yatomi Y, Yazawa M, Di Carlo D, Hosokawa Y, Uemura S, Ozeki Y, Goda K (2018). Intelligent image-activated cell sorting. Cell 175, 266-276. |
| [25] | Ortiz-Ramírez C, Arevalo ED, Xu XS, Jackson DP, Birnbaum KD (2018). An efficient cell sorting protocol for maize protoplasts. Curr Protoc Plant Biol 3, e20072. |
| [26] | Otto FJ (1992). Preparation and staining of cells for high- resolution DNA analysis. In: Radbruch A, ed. Flow Cytometry and Cell Sorting. Berlin: Springer. pp. 65-68. |
| [27] | Petrovská B, Jeřábková H, Chamrád I, Vrána J, Lenobel R, Uřinovská J, Šebela M, Doležel J (2014). Proteomic analysis of barley cell nuclei purified by flow sorting. Cytogenet Genome Res 143, 78-86. |
| [28] | Pfosser M, Heberle-Bors E, Amon A, Lelley T (1995). Evaluation of sensitivity of flow cytometry in detecting aneuploidy in wheat using disomic and ditelosomic wheat- rye addition lines. Cytometry 21, 387-393. |
| [29] | Reichard A, Asosingh K (2019). Best practices for preparing a single cell suspension from solid tissues for flow cytometry. Cytometry A 95, 219-226. |
| [30] | Šafář J, Bartoš J, Janda J, Bellec A, Kubaláková M, Valárik M, Pateyron S, Weiserová J, Tušková R, Číhalíková J, Vrána J, Šimková H, Faivre-Rampant P, Sourdille P, Caboche M, Bernard M, Doležel J, Chalhoub B (2004). Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J 39, 960-968. |
| [31] | Said M, Kubaláková M, Karafiátová M, Molnár I, Doležel J, Vrána J (2019). Dissecting the complex genome of crested wheatgrass by chromosome flow sorting. Plant Genome 12, 180096. |
| [32] | Schraivogel D, Kuhn TM, Rauscher B, Rodríguez-Martínez M, Paulsen M, Owsley K, Middlebrook A, Tischer C, Ramasz B, Ordoñez-Rueda D, Dees M, Cuylen- Haering S, Diebold E, Steinmetz LM (2022). High-speed fluorescence image-enabled cell sorting. Science 375, 315- 320. |
| [33] | Shapiro HM (2003). Practical Flow Cytometry, 4th edn. New York: Wiley-Liss. pp. 73-100. |
| [34] | Sliwinska E, Loureiro J, Leitch IJ, Šmarda P, Bainard J, Bureš P, Chumová Z, Horová L, Koutecký P, Lučanová M, Trávníček P, Galbraith DW (2022). Application-based guidelines for best practices in plant flow cytometry. Cytometry A 101, 749-781. |
| [35] | Smith JP, Sheffield NC (2020). Analytical approaches for ATAC-seq data analysis. Curr Protoc Hum Genet 106, e101. |
| [36] | Sun GL, Xia MZ, Li JP, Ma W, Li QZ, Xie JJ, Bai SL, Fang SS, Sun T, Feng XL, Guo GH, Niu YL, Hou JY, Ye WL, Ma JC, Guo SY, Wang HL, Long Y, Zhang XB, Zhang JL, Zhou H, Li BZ, Liu J, Zou CS, Wang H, Huang JL, Galbraith DW, Song CP (2022). The maize single-nucleus transcriptome comprehensively describes signaling networks governing movement and development of grass stomata. Plant Cell 34, 1890-1911. |
| [37] | Taher TEI (2017). Monitoring promoter activity by flow cytometry. In: Gould D, ed. Mammalian Synthetic Promoters. New York: Humana. pp. 65-73. |
| [38] | The International Wheat Genome Sequencing Consortium IWGSC (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. |
| [39] | Xu XS, Crow M, Rice BR, Li F, Harris B, Liu L, Demesa-Arevalo E, Lu ZF, Wang LY, Fox N, Wang XF, Drenkow J, Luo AD, Char SN, Yang B, Sylvester AW, Gingeras TR, Schmitz RJ, Ware D, Lipka AE, Gillis J, Jackson D (2021). Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev Cell 56, 557-568. |
| [40] | Zhang JD, Feng M (2023). A plant sample optimal pretreatment for flow cytometric analysis. Chin Bull Bot 58, 285-297. (in Chinese) |
| 张晋丹, 冯旻 (2023). 一种提升流式细胞术分析效果的前处理方法. 植物学报 58, 285-297. | |
| [41] | Zhu T, Liao KY, Zhou RF, Xia CJ, Xie WB (2020). ATAC- seq with unique molecular identifiers improves quantification and footprinting. Commun Biol 3, 675. |
| [42] | Zhu T, Xia CJ, Yu RR, Zhou XK, Xu XB, Wang L, Zong ZX, Yang JJ, Liu YM, Ming LC, You YX, Chen DJ, Xie WB (2024). Comprehensive mapping and modelling of the rice regulome landscape unveils the regulatory architecture underlying complex traits. Nat Commun 15, 6562. |