lncRNA调控玉米生长发育和非生物胁迫研究进展
- 中国农业科学院作物科学研究所, 北京 100081
收稿日期: 2024-05-13
录用日期: 2024-07-23
网络出版日期: 2024-08-08
基金资助
国家自然科学基金(32202595)
Research Progress in the Regulation of Development and Stress Responses by Long Non-coding RNAs in Maize
- Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
Received date: 2024-05-13
Accepted date: 2024-07-23
Online published: 2024-08-08
摘要
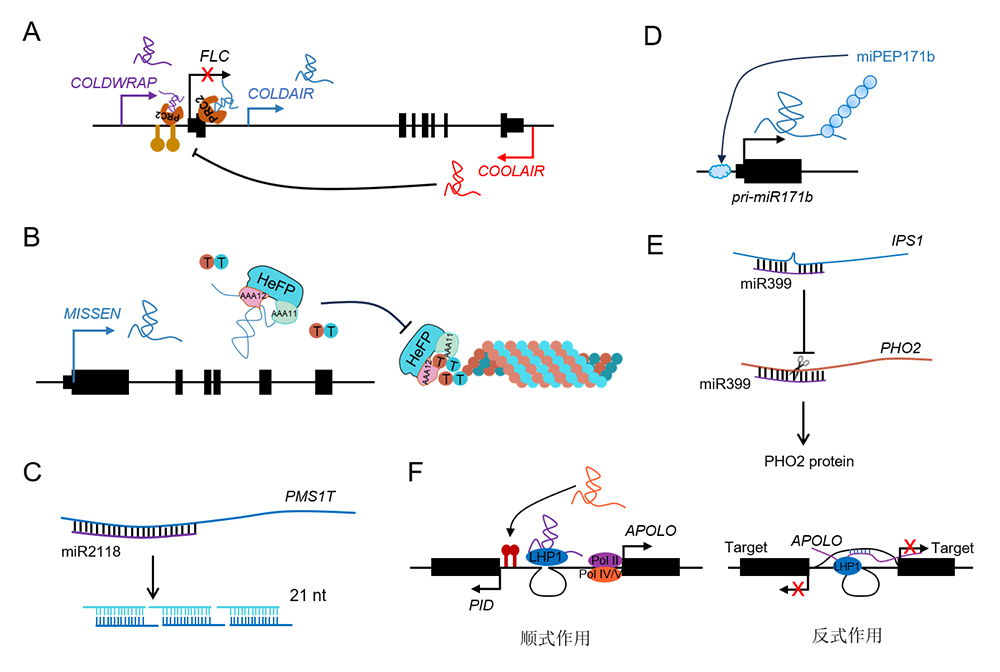
长链非编码RNA (long non-coding RNA, lncRNA)广泛存在于真核生物基因组中, 在维持生物体正常生命活动中发挥重要作用。近年来, 通过高通量测序和生物信息学分析在植物中发掘到大量的lncRNA。已有研究证实lncRNA在调控植物生长发育和逆境响应中发挥重要作用。由于基因组复杂且遗传操作过程繁琐, lncRNA在玉米(Zea mays)中的研究远落后于拟南芥(Arabidopsis thaliana)和水稻(Oryza sativa)。玉米作为我国主要粮食作物, 对于保障国家粮食安全至关重要。玉米还是遗传学与基因组学领域重要的模式植物。了解lncRNA在玉米中的研究进展有助于理解lncRNA的生物学功能。挖掘并解析lncRNA参与玉米生长发育和逆境响应的分子调控网络, 可为玉米遗传改良提供新的分子靶点。该文总结了lncRNA的来源、分类和作用机制, 并讨论了玉米中lncRNA的发掘及其在调控生长发育和逆境响应中的生物学功能, 最后展望了lncRNA在玉米中的研究方向。
本文引用格式
杜庆国 , 李文学 . lncRNA调控玉米生长发育和非生物胁迫研究进展[J]. 植物学报, 2024 , 59(6) : 950 -962 . DOI: 10.11983/CBB24075
Abstract
Long non-coding RNAs (lncRNAs) are widely present in the genomes of eukaryotic organisms and play crucial roles in maintaining the biological activities of living organisms. In recent years, a large number of lncRNAs have been discovered in plants through high-throughput sequencing and bioinformatics analysis. LncRNAs play important roles in regulating plant growth, development, and stress responses. Due to the complexity of the genome and the low efficient of genetic transformation, the functional analysis of lncRNA in maize is relatively lagging behind comparing with that in Arabidopsis and rice. Maize is a major staple crop in China, playing a critical role in ensuring national food security. It’s also an important model plant in the fields of genetics and genomics. Understanding the research progress of lncRNA in maize is highly beneficial for comprehending the biological functions of lncRNA. Mining and analyzing the molecular regulatory network of lncRNAs involved in maize development and stress response can provide potential molecular targets for future genetic improvement of maize. In this review, we summarized the sources, classification, and action mechanisms of lncRNAs, and reviewed the discovery of lncRNAs in maize and their biological functions in regulating growth, development, and stress responses. We also discussed the current research status and provided an outlook on future researches of lncRNAs in maize.

Key words: maize; lncRNA; development; abiotic stress
参考文献
| [1] | Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M (2014). Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell 55, 383-396. |
| [2] | Ariel F, Lucero L, Christ A, Mammarella MF, Jegu T, Veluchamy A, Mariappan K, Latrasse D, Blein T, Liu C, Benhamed M, Crespi M (2020). R-Loop mediated trans action of the APOLO long noncoding RNA. Mol Cell 77, 1055-1065. |
| [3] | Ariel F, Romero-Barrios N, Jégu T, Benhamed M, Crespi M (2015). Battles and hijacks: noncoding transcription in plants. Trends Plant Sci 20, 362-371. |
| [4] | Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141, 1000-1011. |
| [5] | Beló A, Beatty MK, Hondred D, Fengler KA, Li BL, Rafalski A (2010). Allelic genome structural variations in maize detected by array comparative genome hybridization. Theor Appl Genet 120, 355-367. |
| [6] | Boerner S, McGinnis KM (2012). Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS One 7, e43047. |
| [7] | Brannan CI, Dees EC, Ingram RS, Tilghman SM (1990). The product of the H19gene may function as an RNA. Mol Cell Biol 10, 28-36. |
| [8] | Chen DJ, Yuan CH, Zhang J, Zhang Z, Bai L, Meng YJ, Chen LL, Chen M (2012). PlantNATsDB: a comprehensive database of plant natural antisense transcripts. Nucleic Acids Res 40, D1187-D1193. |
| [9] | Chen Q, Zhang XD, Shi JC, Yan MH, Zhou T (2021). Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem Sci 46, 790-804. |
| [10] | Chen XM, Rechavi O (2022). Plant and animal small RNA communications between cells and organisms. Nat Rev Mol Cell Biol 23, 185-203. |
| [11] | Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18, 412-421. |
| [12] | Csorba T, Questa JI, Sun QW, Dean C (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA 111, 16160-16165. |
| [13] | Di Marsico M, Paytuvi-Gallart A, Sanseverino W, Aiese Cigliano R (2022). GreeNC 2.0: a comprehensive database of plant long non-coding RNAs. Nucleic Acids Res 50, D1442-D1447. |
| [14] | Ding JH, Lu Q, Ouyang YD, Mao HL, Zhang PB, Yao JL, Xu CG, Li XH, Xiao JH, Zhang QF (2012). A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci USA 109, 2654-2659. |
| [15] | Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue CH, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, R?der M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang HE, Wrobel J, Yu YB, Ruan XA, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan YJ, Wold B, Carninci P, Guigó R, Gingeras TR (2012). Landscape of transcription in human cells. Nature 489, 101-108. |
| [16] | Downes BP, Stupar RM, Gingerich DJ, Vierstra RD (2003). The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J 35, 729-742. |
| [17] | Du QG, Wang K, Zou C, Xu C, Li WX (2018). The PILNCR1-miR399 regulatory module is important for low phosphate tolerance in maize. Plant Physiol 177, 1743-1753. |
| [18] | Du Toit A (2013). Non-coding RNA: RNA stability control by Pol II. Nat Rev Mol Cell Biol 14, 128-129. |
| [19] | Fan YR, Yang JY, Mathioni SM, Yu JS, Shen JQ, Yang XF, Wang L, Zhang QH, Cai ZX, Xu CG, Li XH, Xiao JH, Meyers BC, Zhang QF (2016). PMS1T, producing phased small-interfering RNAs, regulates photoperiod- sensitive male sterility in rice. Proc Natl Acad Sci USA 113, 15144-15149. |
| [20] | Fang J, Zhang FT, Wang HR, Wang W, Zhao F, Li ZJ, Sun CH, Chen FM, Xu F, Chang SQ, Wu L, Bu QY, Wang PR, Xie JK, Chen F, Huang XH, Zhang YJ, Zhu XG, Han B, Deng XJ, Chu CC (2019). Ef-cd locus shortens rice maturity duration without yield penalty. Proc Natl Acad Sci USA 116, 18717-18722. |
| [21] | Fang XF, Wu Z, Raitskin O, Webb K, Voigt P, Lu TC, Howard M, Dean C (2020). The 3' processing of antisense RNAs physically links to chromatin-based transcriptional control. Proc Natl Acad Sci USA 117, 15316-15321. |
| [22] | Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39, 1033-1037. |
| [23] | Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15, 2038-2043. |
| [24] | Guo GH, Liu XY, Sun FL, Cao J, Huo N, Wuda B, Xin MM, Hu ZR, Du JK, Xia R, Rossi V, Peng HR, Ni ZF, Sun QX, Yao YY (2018). Wheat miR9678 affects seed germination by generating phased siRNAs and modulating abscisic acid/gibberellin signaling. Plant Cell 30, 796-814. |
| [25] | Han LQ, Mu ZN, Luo Z, Pan QC, Li L (2019). New lncRNA annotation reveals extensive functional divergence of the transcriptome in maize. J Integr Plant Biol 61, 394-405. |
| [26] | He ZH, Lan YM, Zhou XK, Yu BJ, Zhu T, Yang F, Fu LY, Chao HY, Wang JH, Feng RX, Zuo SM, Lan WZ, Chen CL, Chen M, Zhao X, Hu KM, Chen DJ (2024). Single- cell transcriptome analysis dissects lncRNA-associated gene networks in Arabidopsis. Plant Commun 5, 100717. |
| [27] | Heo JB, Sung S (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76-79. |
| [28] | Hermans-Beijnsberger S, van Bilsen M, Schroen B (2018). Long non-coding RNAs in the failing heart and vasculature. Noncoding RNA Res 3, 118-130. |
| [29] | Hong YH, Zhang YY, Cui J, Meng J, Chen YH, Zhang CW, Yang JX, Luan YS (2022). The lncRNA39896-miR166b- HDZs module affects tomato resistance to Phytophthora infestans. J Integr Plant Biol 64, 1979-1993. |
| [30] | Hu B, Zhu CG, Li F, Tang JY, Wang YQ, Lin AH, Liu LC, Che RH, Chu CC (2011). LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol 156, 1101-1115. |
| [31] | Hu XL, Wei QY, Wu HY, Huang YX, Peng XJ, Han GM, Ma Q, Zhao Y (2022). Identification and characterization of heat-responsive lncRNAs in maize inbred line CM1. BMC Genomics 23, 208. |
| [32] | Huang DQ, Feurtado JA, Smith MA, Flatman LK, Koh C, Cutler AJ (2017). Long noncoding miRNA gene represses wheat β-diketone waxes. Proc Natl Acad Sci USA 114, E3149-E3158. |
| [33] | Huang TK, Han CL, Lin SI, Chen YJ, Tsai YC, Chen YR, Chen JW, Lin WY, Chen PM, Liu TY, Chen YS, Sun CM, Chiou TJ (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25, 4044-4060. |
| [34] | Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu QY, Poirier Y (2013). A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25, 4166-4182. |
| [35] | Jin JJ, Lu P, Xu YL, Li ZF, Yu SZ, Liu J, Wang H, Chua NH, Cao PJ (2021). PLncDB V2.0: a comprehensive encyclopedia of plant long noncoding RNAs. Nucleic Acids Res 49, D1489-D1495. |
| [36] | Kim DH, Sung S (2017). Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev Cell 40, 302-312. |
| [37] | Lai JS, Li RQ, Xu X, Jin WW, Xu ML, Zhao HN, Xiang ZK, Song WB, Ying K, Zhang M, Jiao YP, Ni PX, Zhang JG, Li D, Guo XS, Ye KX, Jian M, Wang B, Zheng HS, Liang HQ, Zhang XQ, Wang SC, Chen SJ, Li JS, Fu Y, Springer NM, Yang HM, Wang J, Dai JR, Schnable PS, Wang J (2010). Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42, 1027-1030. |
| [38] | Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, Bécard G, Combier JP (2015). Primary transcripts of microRNAs encode regulatory peptides. Nature 520, 90-93. |
| [39] | Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, Evans MMS, Scanlon MJ, Yu JM, Schnable PS, Timmermans MCP, Springer NM, Muehlbauer GJ (2014). Genome- wide discovery and characterization of maize long non- coding RNAs. Genome Biol 15, R40. |
| [40] | Li W, Chen YD, Wang YL, Zhao J, Wang YJ (2022a). Gypsy retrotransposon-derived maize lncRNA GARR2 modulates gibberellin response. Plant J 110, 1433-1446. |
| [41] | Li XH, Chen WW, Lu SQ, Fang JT, Zhu H, Zhang XB, Qi YW (2022b). Full-length transcriptome analysis of maize root tips reveals the molecular mechanism of cold stress during the seedling stage. BMC Plant Biol 22, 398. |
| [42] | Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147, 732-746. |
| [43] | Liu P, Zhang YC, Zou CY, Yang C, Pan GT, Ma LL, Shen YO (2022). Integrated analysis of long non-coding RNAs and mRNAs reveals the regulatory network of maize seedling root responding to salt stress. BMC Genomics 23, 50. |
| [44] | Lou DJ, Li F, Ge JY, Fan WY, Liu ZR, Wang YY, Huang JF, Xing M, Guo WL, Wang SZ, Qiao WH, Han ZY, Qian Q, Yang QW, Zheng XM (2022). LncPheDB: a genome- wide lncRNAs regulated phenotypes database in plants. aBIOTECH 3, 169-177. |
| [45] | Lu J, Zhen S, Zhang J, Xie Y, He C, Wang X, Wang Z, Zhang S, Li Y, Cui Y, Wang G, Wang J, Liu J, Li L, Gu R, Zheng X, Fu J (2023). Combined population transcriptomic and genomic analysis reveals cis-regulatory differentiation of non-coding RNAs in maize. Theor Appl Genet 136, 16. |
| [46] | Lv YD, Liang ZK, Ge M, Qi WC, Zhang TF, Lin F, Peng ZH, Zhao H (2016). Genome-wide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize (Zea mays L.). BMC Genomics 17, 350. |
| [47] | Ma JX, Yan BX, Qu YY, Qin FF, Yang YT, Hao XJ, Yu JJ, Zhao Q, Zhu DY, Ao GM (2008). Zm401, a short-open reading-frame mRNA or noncoding RNA, is essential for tapetum and microspore development and can regulate the floret formation in maize. J Cell Biochem 105, 136-146. |
| [48] | Mao Y, Xu J, Wang Q, Li GB, Tang X, Liu TH, Feng XJ, Wu FK, Li ML, Xie WB, Lu YL (2021). A natural antisense transcript acts as a negative regulator for the maize drought stress response gene ZmNAC48. J Exp Bot 72, 2790-2806. |
| [49] | Melé M, Mattioli K, Mallard W, Shechner DM, Gerhardinger C, Rinn JL (2017). Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res 27, 27-37. |
| [50] | Moison M, Pacheco JM, Lucero L, Fonouni-Farde C, Rodríguez-Melo J, Mansilla N, Christ A, Bazin J, Benhamed M, Iba?ez F, Crespi M, Estevez JM, Ariel F (2021). The lncRNA APOLO interacts with the transcription factor WRKY42 to trigger root hair cell expansion in response to cold. Mol Plant 14, 937-948. |
| [51] | Ng SY, Lin L, Soh BS, Stanton LW (2013). Long noncoding RNAs in development and disease of the central nervous system. Trends Genet 29, 461-468. |
| [52] | Palazzo AF, Koonin EV (2020). Functional long non-coding RNAs evolve from junk transcripts. Cell 183, 1151-1161. |
| [53] | Ponting CP, Oliver PL, Reik W (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629-641. |
| [54] | Qin T, Zhao HY, Cui P, Albesher N, Xiong LM (2017). A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol 175, 1321-1336. |
| [55] | Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159-1169. |
| [56] | R?hrig H, Schmidt J, Miklashevichs E, Schell J, John M (2002). Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc Natl Acad Sci USA 99, 1915-1920. |
| [57] | Sharma A, Badola PK, Bhatia C, Sharma D, Trivedi PK (2020). Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis. Nat Plants 6, 1262-1274. |
| [58] | Shi XF, Sun M, Liu HB, Yao YW, Song Y (2013). Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 339, 159-166. |
| [59] | Song YP, Xuan AR, Bu CH, Ci D, Tian M, Zhang DQ (2019). Osmotic stress-responsive promoter upstream transcripts (PROMPTs) act as carriers of MYB transcription factors to induce the expression of target genes in Populus simonii. Plant Biotechnol J 17, 164-177. |
| [60] | Statello L, Guo CJ, Chen LL, Huarte M (2020). Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22, 96-118. |
| [61] | Sun Q, Liu XG, Yang J, Liu WW, Du QG, Wang HQ, Fu CX, Li WX (2018). MicroRNA528 affects lodging resistance of maize by regulating lignin biosynthesis under nitrogen-luxury conditions. Mol Plant 11, 806-814. |
| [62] | Tadege M, Sheldon CC, Helliwell CA, Upadhyaya NM, Dennis ES, Peacock WJ (2003). Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol J 1, 361-369. |
| [63] | Tang X, Li QM, Feng XJ, Yang B, Zhong X, Zhou Y, Wang Q, Mao Y, Xie WB, Liu TH, Tang Q, Guo W, Wu FK, Feng XJ, Wang QJ, Lu YL, Xu J (2023). Identification and functional analysis of drought-responsive long noncoding RNAs in maize roots. Int J Mol Sci 24, 15039. |
| [64] | Tian YK, Zheng H, Zhang F, Wang SL, Ji XR, Xu C, He YH, Ding Y (2019). PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci Adv 5, eaau7246. |
| [65] | Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6, e1001031. |
| [66] | Wang H, Niu QW, Wu HW, Liu J, Ye J, Yu N, Chua NH (2015). Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J 84, 404-416. |
| [67] | Wang KC, Chang HY (2011). Molecular mechanisms of long noncoding RNAs. Mol Cell 43, 904-914. |
| [68] | Wang TZ, Zhao MG, Zhang XX, Liu M, Yang CG, Chen YH, Chen RJ, Wen JQ, Mysore KS, Zhang WH (2017). Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J Exp Bot 68, 5937-5948. |
| [69] | Wang Y, Luo XJ, Sun F, Hu JH, Zha X, Su W, Yang JS (2018). Overexpressing lncRNA LAIR increases grain yield and regulates neighbouring gene cluster expression in rice. Nat Commun 9, 3516. |
| [70] | Wang YF, Wang ZH, Du QG, Wang K, Zou CQ, Li WX (2023). The long non-coding RNA PILNCR2 increases low phosphate tolerance in maize by interfering with miRNA399-guided cleavage of ZmPHT1s. Mol Plant 16, 1146-1159. |
| [71] | Wang YQ, Fan XD, Lin F, He GM, Terzaghi W, Zhu DM, Deng XW (2014). Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci USA 111, 10359-10364. |
| [72] | Wierzbicki AT, Haag JR, Pikaard CS (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635-648. |
| [73] | Xing LJ, Zhu M, Luan MD, Zhang M, Jin L, Liu YP, Zou JJ, Wang L, Xu MY (2022). miR169q and NUCLEAR FACTOR YA8 enhance salt tolerance by activating PEROXIDASE1 expression in response to ROS. Plant Physiol 188, 608-623. |
| [74] | Yang J, Wei HB, Hou M, Chen LH, Zou T, Ding H, Jing YF, Zhang XM, Zhao YP, Liu Q, Heng YQ, Wu H, Wang BB, Kong DX, Wang HY (2023). ZmSPL13 and ZmSPL29 act together to promote vegetative and reproductive transition in maize. New Phytol 239, 1505-1520. |
| [75] | Yi X, Zhang ZH, Ling Y, Xu WY, Su Z (2015). PNRD: a plant non-coding RNA database. Nucleic Acids Res 43, D982-D989. |
| [76] | Zhan JP, Meyers BC (2023). Plant small RNAs: their biogenesis, regulatory roles, and functions. Annu Rev Plant Biol 74, 21-51. |
| [77] | Zhang N, Liu ZG, Sun SC, Liu SY, Lin JH, Peng YF, Zhang XX, Yang H, Cen X, Wu J (2020). Response of AtR8 lncRNA to salt stress and its regulation on seed germination in Arabidopsis. Chin Bull Bot 55, 421-429. (in Chinese) |
| 张楠, 刘自广, 孙世臣, 刘圣怡, 林建辉, 彭疑芳, 张晓旭, 杨贺, 岑曦, 吴娟 (2020). 拟南芥AtR8 IncRNA对盐胁迫响应及其对种子萌发的调节作用. 植物学报 55, 421-429. | |
| [78] | Zhang S, Wu CY (2019). Long noncoding RNA Ef-cd promotes maturity without yield penalty in rice. Chin Bull Bot 54, 550-553. (in Chinese) |
| 张硕, 吴昌银 (2019). 长链非编码RNA基因Ef-cd调控水稻早熟与稳产. 植物学报 54, 550-553. | |
| [79] | Zhang ZF, Xu Y, Yang F, Xiao BZ, Li GL (2021). RiceLncPedia: a comprehensive database of rice long non- coding RNAs. Plant Biotechnol J 19, 1492-1494. |
| [80] | Zhang ZH, Zhang RJ, Meng FF, Chen YM, Wang WX, Yang K, Gao YJ, Xin MM, Du JK, Hu ZR, Ni ZF, Sun QX, Guo WL, Yao YY, Zhang ZH, Zhang RJ, Meng FF, Chen YM, Wang WX, Yang K, Gao YJ, Xin MM, Du JK, Hu ZR, Ni ZF, Sun QX, Guo WL, Yao YY (2023). A comprehensive atlas of long non-coding RNAs provides insight into grain development in wheat. Seed Biol 2, 12. |
| [81] | Zhao SR, Zhang Y, Gordon W, Quan J, Xi HL, Du S, von Schack D, Zhang BH (2015). Comparison of stranded and non-stranded RNA-seq transcriptome profiling and investigation of gene overlap. BMC Genomics 16, 675. |
| [82] | Zhao XY, Li JR, Lian B, Gu HQ, Li Y, Qi YJ (2018). Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat Commun 9, 5056. |
| [83] | Zhao YS, Zhu P, Hepworth J, Bloomer R, Antoniou- Kourounioti RL, Doughty J, Heckmann A, Xu CY, Yang HC, Dean C (2021). Natural temperature fluctuations promote COOLAIR regulation of FLC. Genes Dev 35, 888-898. |
| [84] | Zhou YF, Zhang YC, Sun YM, Yu Y, Lei MQ, Yang YW, Lian JP, Feng YZ, Zhang Z, Yang L, He RR, Huang JH, Cheng Y, Liu YW, Chen YQ (2021). The parent-of-origin lncRNA MISSEN regulates rice endosperm development. Nat Commun 12, 6525. |