

水稻“混血杂交”群体揭示遗传互作奥秘
收稿日期: 2024-06-07
录用日期: 2024-06-21
网络出版日期: 2024-07-05
基金资助
国家自然科学基金委区域联合基金(U22A20465)
The Rice "hybrid" Population Reveals the Mysteries of Genetic Interaction
Received date: 2024-06-07
Accepted date: 2024-06-21
Online published: 2024-07-05
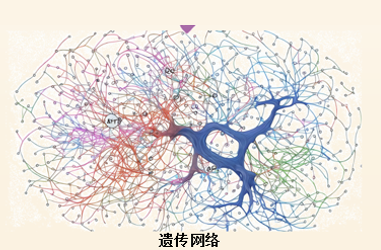
基因互作与表型的内在关系是生命科学研究的关键问题, 大部分表型受多基因协同控制, 除加性效应外还存在显性和上位性等复杂遗传效应。最近一项研究构建了包含18 421个永久株系的水稻“混血杂交”群体, 成功鉴定到控制16个农艺性状的96个高置信候选基因, 通过分析基因间上位性效应, 构建了包含19个枢纽基因的遗传互作网络, 揭示出基因间潜在的互作效应, 发现170个“掩蔽”型上位互作对。该工作建立了作物遗传学研究的新范式, 且为水稻(Oryza sativa)遗传研究提供了重要数据和材料资源, 极大地加速了重要性状相关基因的挖掘, 推动了数量性状基因遗传互作的功能解析, 为分子设计育种奠定了理论基础。
王淏 , 钦鹏 , 李仕贵 . 水稻“混血杂交”群体揭示遗传互作奥秘[J]. 植物学报, 2024 , 59(4) : 529 -532 . DOI: 10.11983/CBB24090
Revealing the intrinsic relationship between gene interactions and phenotypes is a key issue in life science research. Most phenotypes are controlled by multiple genes in a coordinated manner, exhibiting complex genetic effects such as dominance and epistasis in addition to additive effects. In a recent study, a “hybrid” population of rice containing 18 421 recombinant inbred lines was constructed, successfully identifying 96 high-confidence candidate genes controlling 16 agronomic traits. The study analyzed the epistatic effects among genes, constructed a genetic interaction network consisting of 19 hub genes, revealed potential interactions among genes, and discovered 170 “masking” epistatic interac-tions. This research provides important data and material resources for genetic studies in rice, establishes a new paradigm for crop genetics research, greatly accelerates the discovery of genes related to important traits, advances the functional analysis of quantitative trait gene interactions, and provides genetic resources and theoretical support for mole-cular breeding.

Key words: “hybrid” population; quantitative traits; genetic pleiotropy; epistasis
| [1] | Forsberg SKG, Bloom JS, Sadhu MJ, Kruglyak L, Carlborg ? (2017). Accounting for genetic interactions improves modeling of individual quantitative trait phenotypes in yeast. Nat Genet 49, 497-503. |
| [2] | Gros PA, Le Nagard H, Tenaillon O (2009). The evolution of epistasis and its links with genetic robustness, complexity and drift in a phenotypic model of adaptation. Genetics 182, 277-293. |
| [3] | Li T, Ning Z, Yang ZJ, Zhai RR, Zheng CQ, Xu WZ, Wang YP, Ying KJ, Chen YW, Shen X (2021). Total genetic contribution assessment across the human genome. Nat Commun 12, 2845. |
| [4] | Liu HJ, Yan JB (2019). Crop genome-wide association study: a harvest of biological relevance. Plant J 97, 8-18. |
| [5] | Liu YQ, Wang HR, Jiang ZM, Wang W, Xu RN, Wang QH, Zhang ZH, Li AF, Liang Y, Ou SJ, Liu XJ, Cao SY, Tong HN, Wang YH, Zhou F, Liao H, Hu B, Chu CC (2021). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590, 600-605. |
| [6] | Si LZ, Chen JY, Huang XH, Gong H, Luo JH, Hou QQ, Zhou TY, Lu TT, Zhu JJ, Shangguan YY, Chen EW, Gong CX, Zhao Q, Jing YF, Zhao Y, Li Y, Cui LL, Fan DL, Lu YQ, Weng QJ, Wang YC, Zhan QL, Liu KY, Wei XH, An K, An G, Han B (2016). OsSPL13 controls grain size in cultivated rice. Nat Genet 48, 447-456. |
| [7] | Stearns FW (2010). One hundred years of pleiotropy: a retrospective. Genetics 186, 767-773. |
| [8] | Watanabe K, Stringer S, Frei O, Umi?evi? Mirkov M, de Leeuw C, Polderman TJC, van der Sluis S, Andreassen OA, Neale BM, Posthuma D (2019). A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet 51, 1339-1348. |
| [9] | Wei X, Chen M, Zhang Q, Gong J, Liu J, Yong K, Wang Q, Fan J, Chen S, Hua H, Luo Z, Zhao X, Wang X, Li W, Cong J, Yu X, Wang Z, Huang R, Chen J, Zhou X, Qiu J, Xu P, Murray J, Wang H, Xu Y, Xu C, Xu G, Yang J, Han B, Huang X (2024). Genomic investigation of 18 421 lines reveals the genetic architecture of rice. Science 385, eadm8762. |
/
| 〈 |
|
〉 |