

AtGH3.17调控拟南芥生长素和油菜素甾醇的响应
收稿日期: 2022-04-05
录用日期: 2022-07-25
网络出版日期: 2022-07-27
基金资助
国家自然科学基金(31900176);广东省自然科学基金(2020A1515011387)
AtGH3.17-mediated Regulation of Auxin and Brassinosteroid Response in Arabidopsis thaliana
Received date: 2022-04-05
Accepted date: 2022-07-25
Online published: 2022-07-27
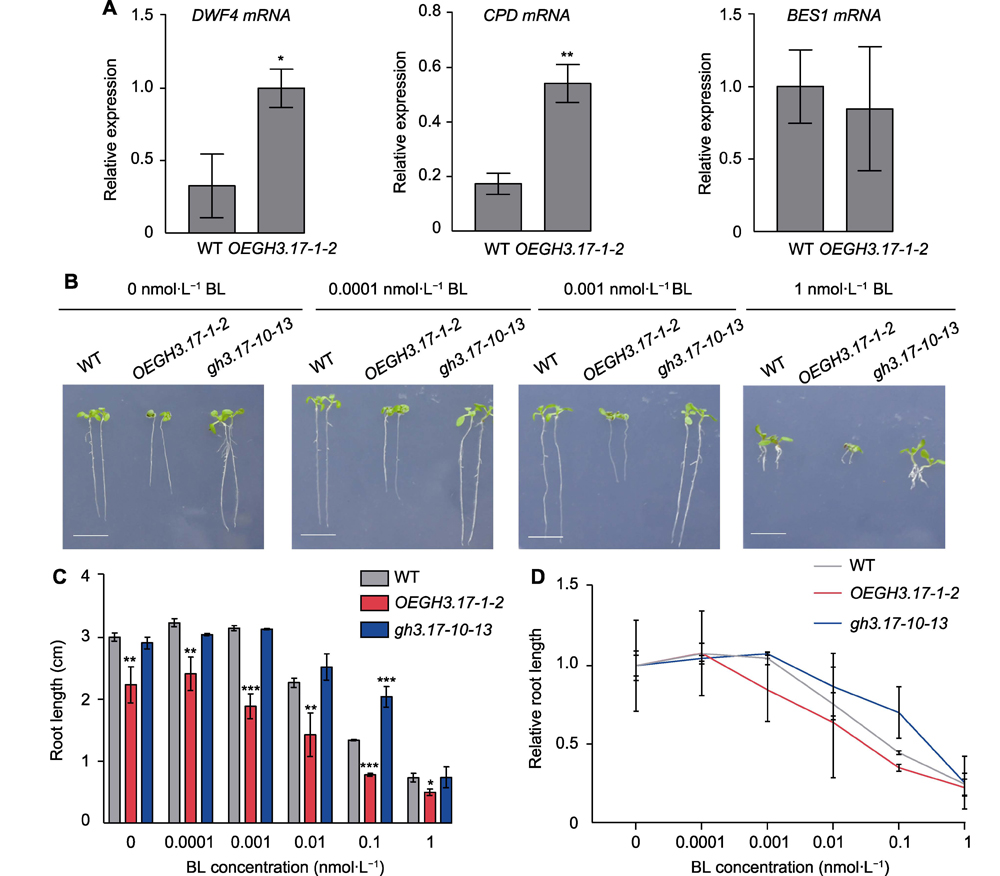
植物激素油菜素甾醇和生长素在植物生长发育过程中发挥重要作用, 且两者之间存在复杂的调控关系。为探究拟南芥(Arabidopsis thaliana)生长素早期响应基因GH3.17是否参与生长素和油菜素甾醇之间的调控, 利用转基因技术和CRISPR/Cas9基因编辑系统分别获得拟南芥GH3.17过表达和缺失突变植株, 并对其表型进行比较分析。结果显示, 与野生型相比, GH3.17缺失突变体的根长增加, 而GH3.17过表达植株呈现根长变短、叶片卷曲、叶柄缩短和株高变矮等典型的生长素缺陷表型。采用一定浓度的生长素或者油菜素甾醇处理植株, 结果显示过表达植株的根明显变长, 表明其对生长素以及油菜素甾醇的响应更为敏感。通过qRT-PCR分析过表达植株中生长素信号通路和油菜素甾醇合成通路相关基因的表达情况, 发现生长素信号通路基因Aux/IAA家族成员IAA12和IAA16的表达受到抑制, 而油菜素甾醇合成酶基因DWF4和CPD的表达量增高。综上, 推测活性生长素含量减少导致的生长素信号被抑制可以通过增加油菜素甾醇合成来进行一定的补偿。
周淑瑶 , 李建明 , 毛娟 . AtGH3.17调控拟南芥生长素和油菜素甾醇的响应[J]. 植物学报, 2023 , 58(3) : 373 -384 . DOI: 10.11983/CBB22063
Brassinosteroid and auxin play important roles in plant growth and development. There is a complex crosstalk between the two plant hormones. In order to study whether GH3.17 is involved in the crosstalk between auxin and brassinosteroid (BR), we created overexpression and CRISPR gene loss mutants of Arabidopsis GH3.17. The root length of GH3.17 CRISPR mutants was slightly longer than that of wild-type plants. However, the GH3.17 overexpression seedlings had typical auxin deficiency phenotypes with decreased height, curly leaves and shorten roots and petioles. The root length of GH3.17 overexpression seedlings was substantially increased by the application of low concentrations of indole acetic acid (IAA) or brassinolide (BL), and the overexpression plants were more sensitive to IAA and BR than the wild type plants. We also found that the expression of auxin signaling, and BR synthesis pathway-related genes IAA12 and IAA16 was inhibited in the overexpression plants, while the expression abundance of brassinosteroid biosynthetic genes DWF4 and CPD was increased, suggesting that the inhibition of auxin signal transduction caused by the deficiency of active auxin may be compensated by BR compounds accumulation.

Key words: auxin; brassinosteroid; early auxin-response gene; GH3.17
| [1] | 李艳艳, 齐艳华 (2022). 植物Aux/IAA基因家族生物学功能研究进展. 植物学报 57, 30-41. |
| [2] | 任鸿雁, 王莉, 马青秀, 吴光 (2015). 油菜素内酯生物合成途径的研究进展. 植物学报 50, 768-778. |
| [3] | 孙超, 黎家 (2017). 油菜素甾醇类激素的生物合成、代谢及信号转导. 植物生理学报 53, 291-307. |
| [4] | 王冰, 李家洋, 王永红 (2006). 生长素调控植物株型形成的研究进展. 植物学通报 23, 443-458. |
| [5] | 谢先荣, 曾栋昌, 谭健韬, 祝钦泷, 刘耀光 (2021). 基于CRISPR编辑系统的DNA片段删除技术. 植物学报 56, 44-49. |
| [6] | Abel S, Theologis A (1996). Early genes and auxin action. Plant Physiol 111, 9-17. |
| [7] | Bao F, Shen JJ, Brady SR, Muday GK, Asami T, Yang ZB (2004). Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 134, 1624-1631. |
| [8] | Chung Y, Maharjan PM, Lee O, Fujioka S, Jang S, Kim B, Takatsuto S, Tsujimoto M, Kim H, Cho S, Park T, Cho H, Hwang I, Choe S (2011). Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J 66, 564-578. |
| [9] | Clouse SD, Sasse JM (1998). BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49, 427-451. |
| [10] | Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novak O, Ljung K, Di Paola L, Marée AFM, Costantino P, Grieneisen VA, Sabatini S (2017). Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci USA 114, E7641-E7649. |
| [11] | Di Mambro R, Svolacchia N, Dello Ioio R, Pierdonati E, Salvi E, Pedrazzini E, Vitale A, Perilli S, Sozzani R, Benfey PN, Busch W, Costantino P, Sabatini S (2019). The lateral root cap acts as an auxin sink that controls meristem size. Curr Biol 29, 1199-1205. |
| [12] | Ding ZJ, Friml J (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci USA 107, 12046-12051. |
| [13] | Du MM, Spalding EP, Gray WM (2020). Rapid auxin-mediated cell expansion. Annu Rev Plant Biol 71, 379-402. |
| [14] | Favero DS, Le KN, Neff MM (2017). Brassinosteroid signaling converges with SUPPRESSOR OF PHYTOCHROME B4-#3 to influence the expression of SMALL AUXIN UP RNA genes and hypocotyl growth. Plant J 89, 1133-1145. |
| [15] | Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134, 1555-1573. |
| [16] | Hagen G, Guilfoyle T (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49, 373-385. |
| [17] | Hagen G, Guilfoyle TJ (1985). Rapid induction of selective transcription by auxins. Mol Cell Biol 5, 1197-1203. |
| [18] | Hirano K, Yoshida H, Aya K, Kawamura M, Hayashi M, Hobo T, Sato-Izawa K, Kitano H, Ueguchi-Tanaka M, Matsuoka M (2017). SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING form a complex to integrate auxin and brassinosteroid signaling in rice. Mol Plant 10, 590-604. |
| [19] | Hsieh HL, Okamoto H, Wang ML, Ang LH, Matsui M, Goodman H, Deng XW (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14, 1958-1970. |
| [20] | Iba?es M, Fàbregas N, Chory J, Ca?o-Delgado AI (2009). Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc Natl Acad Sci USA 106, 13630-13635. |
| [21] | Liu XL, Yang HX, Wang Y, Zhu ZH, Zhang W, Li JM (2020). Comparative transcriptomic analysis to identify b- rassinosteroid response genes. Plant Physiol 184, 1072-1082. |
| [22] | Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ (1994). Soybean GH3promoter contains multiple auxin-inducible elements. Plant Cell 6, 645-657. |
| [23] | Luo J, Zhou JJ, Zhang JZ (2018). Aux/IAA gene family in plants: molecular structure, regulation, and function. Int J Mol Sci 19, 259. |
| [24] | Mouchel CF, Osmont KS, Hardtke CS (2006). BRX mediates feedback between brassinosteroid levels and auxin signaling in root growth. Nature 443, 458-461. |
| [25] | Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S (2003). Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133, 1843-1853. |
| [26] | Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25, 213-221. |
| [27] | Ohnishi T, Szatmari AM, Watanabe B, Fujita S, Bancos S, Koncz C, Lafos M, Shibata K, Yokota T, Sakata K, Szekeres M, Mizutani M (2006). C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18, 3275-3288. |
| [28] | Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu GX, Theologis A (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444-463. |
| [29] | Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M (2009). Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106, 22540-22545. |
| [30] | Sakamoto T, Morinaka Y, Inukai Y, Kitano H, Fujioka S (2013). Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J 73, 676-688. |
| [31] | Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616-627. |
| [32] | Suzuki H, Fujioka S, Takatsuto S, Yokota T, Murofushi N, Sakurai A (1994). Biosynthesis of brassinolide from teasterone via typhasterol and castasterone in cultured cells of Catharanthus roseus. J Plant Growth Regul 13, 21-26. |
| [33] | Tan X, Calderon-Villalobos LIA, Sharon M, Zheng CX, Robinson CV, Estelle M, Zheng N (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640-645. |
| [34] | Ulmasov T, Hagen G, Guilfoyle TJ (1997a). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865-1868. |
| [35] | Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995). Composite structure of auxin response elements. Plant Cell 7, 1611-1623. |
| [36] | Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963-1971. |
| [37] | Wang ZY, Bai MY, Oh E, Zhu JY (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46, 701-724. |
| [38] | Weijers D, Benkova E, J?ger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G (2005). Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24, 1874-1885. |
| [39] | Woodward AW, Bartel B (2005). Auxin: regulation, action, and interaction. Ann Bot 95, 707-735. |
| [40] | Yoshimitsu Y, Tanaka K, Fukuda W, Asami T, Yoshida S, Hayashi KI, Kamiya Y, Jikumaru Y, Shigeta T, Nakamura Y, Matsuo T, Okamoto S (2011). Transcription of DWARF4 plays a crucial role in auxin-regulated root elongation in addition to brassinosteroid homeostasis in Arabidopsis thaliana. PLoS One 6, e23851. |
| [41] | Zhang C, Zhang L, Wang D, Ma H, Liu B, Shi Z, Ma X, Chen Y, Chen Q (2018). Evolutionary history of the Glycoside Hydrolase 3 (GH3) family based on the sequenced genomes of 48 plants and identification of jasmonic acid- related GH3 proteins in Solanum tuberosum. Int J Mol Sci 19, 1850. |
| [42] | Zheng ZY, Guo YX, Novák O, Chen W, Ljung K, Noel JP, Chory J (2016). Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat Plants 2, 16025. |
/
| 〈 |
|
〉 |