

植物光信号途径重要新调控因子TZP的研究进展
收稿日期: 2022-04-15
录用日期: 2022-06-23
网络出版日期: 2022-06-23
基金资助
国家自然科学基金(32000187)
Research Progress on TZP, a Novel Key Regulator of Light Signal Transduction in Plants
Received date: 2022-04-15
Accepted date: 2022-06-23
Online published: 2022-06-23
李聪, 齐立娟, 谷晓峰, 李继刚 . 植物光信号途径重要新调控因子TZP的研究进展[J]. 植物学报, 2022 , 57(5) : 579 -587 . DOI: 10.11983/CBB22076
TANDEM ZINC-FINGER/PLUS3 (TZP) is a recently characterized novel key component of light signal transduction in plants. Accumulating evidence indicates that TZP plays an important role in multiple processes of light-mediated plant growth and development. TZP acts as a negative regulator of blue light signaling, regulates phytochrome B (phyB)-dependent control of photoperiodic flowering, and is required for the phosphorylation of the far-red light photoreceptor phyA in vivo. Further investigation and full elucidation of the biochemical activity and regulatory mechanisms of TZP will not only provide new insights into the light signaling networks, but also facilitate the design and breeding of new crop varieties with ideal plant architecture and more efficient usage of solar energy. In this review, we systematically summarized the current understanding of the role of TZP in regulating light signaling pathways in plants, and discussed the important questions for further study on TZP functions.
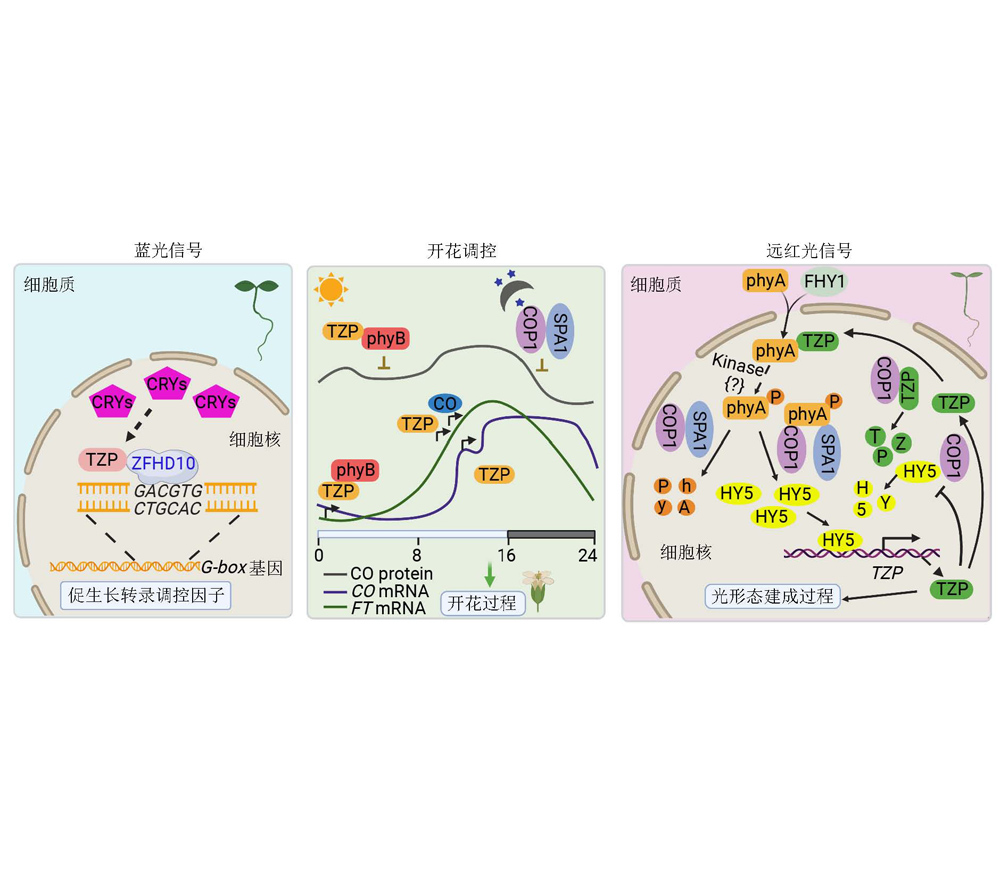
Key words: TZP; light signaling; phytochrome; regulatory mechanism; biochemical function
| [1] | 景艳军, 林荣呈 (2017). 我国植物光信号转导研究进展概述. 植物学报 52, 257-270. |
| [2] | 李聪 (2019). 远红光信号调控因子TZP的作用机制研究. 博士论文. 北京: 中国农业大学. pp. 60-82. |
| [3] | 刘明雪, 孙梅, 王宇, 李玉花 (2012). 植物UV-B受体及其介导的光信号转导. 植物学报 47, 661-669. | [4] | 马朝峰, 戴思兰 (2019). 光受体介导信号转导调控植物开花研究进展. 植物学报 54, 9-22. |
| [5] | 王静, 王艇 (2007). 高等植物光敏色素的分子结构?生理功能和进化特征. 植物学通报 24, 649-658. |
| [6] | 张少蔓 (2019). TZP调控植物远红光信号途径的分子机制研究. 博士论文. 北京: 中国农业大学. pp. 50-66. |
| [7] | 周杨杨, 李继刚 (2019). 植物远红光受体——光敏色素A的研究进展. 自然杂志 41, 188-196. |
| [8] | Bourbousse C, Ahmed I, Roudier F, Zabulon G, Blondet E, Balzergue S, Colot V, Bowler C, Barneche F (2012). Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet 8, e1002825. |
| [9] | Burko Y, Seluzicki A, Zander M, Pedmale UV, Ecker JR, Chory J (2020). Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell 32, 967-983. |
| [10] | Ca?ibano E, Bourbousse C, García-León M, Gómez BG, Wolff L, García-Baudino C, Lozano-Durán R, Barneche F, Rubio V, Fonseca S (2021). DET1-mediated COP1 regulation avoids HY5 activity over second-site gene targets to tune plant photomorphogenesis. Mol Plant 14, 963-982. |
| [11] | Cerdán PD, Chory J (2003). Regulation of flowering time by light quality. Nature 423, 881-885. |
| [12] | Cheng MC, Kathare PK, Paik I, Huq E (2021). Phytochrome signaling networks. Annu Rev Plant Biol 72, 217-244. |
| [13] | Chow BY, Helfer A, Nusinow DA, Kay SA (2012). ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal Behav 7, 170-173. |
| [14] | Clay NK, Nelson T (2005). The recessive epigenetic swellmap mutation affects the expression of two step II splicing factors required for the transcription of the cell proliferation gene STRUWWELPETER and for the timing of cell cycle arrest in the Arabidopsis leaf. Plant Cell 17, 1994-2008. |
| [15] | De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF (1998). Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science 279, 384-388. |
| [16] | De Jong RN, Truffault V, Diercks T, Ab E, Daniels MA, Kaptein R, Folkers GE (2008). Structure and DNA binding of the human Rtf1 Plus3 domain. Structure 16, 149-159. |
| [17] | Han X, Huang X, Deng XW (2020). The photomorphogenic central repressor COP1: conservation and functional diversification during evolution. Plant Commun 1, 100044. |
| [18] | Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21, 126-133. |
| [19] | Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang MM, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, Webb A, Gon?alves J, Davis SJ (2012). EARLY FLOWERING 4 recruitment of EARLY FLOWERING 3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24, 428-443. |
| [20] | Hiltbrunner A, Viczián A, Bury E, Tscheuschler A, Kircher S, Tóth R, Honsberger A, Nagy F, Fankhauser C, Sch?fer E (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol 15, 2125-2130. |
| [21] | Huang H, Alvarez S, Bindbeutel R, Shen ZX, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, Nusinow DA (2016). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol Cell Proteomics 15, 201-217. |
| [22] | Hudson M, Ringli C, Boylan MT, Quail PH (1999). The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13, 2017-2027. |
| [23] | Jiao YL, Lau OS, Deng XW (2007). Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8, 217-230. |
| [24] | Kaiserli E, Páldi K, O'Donnell L, Batalov O, Pedmale UV, Nusinow DA, Kay SA, Chory J (2015). Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev Cell 35, 311-321. |
| [25] | Kalyna M, Lopato S, Barta A (2003). Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol Biol Cell 14, 3565-3577. |
| [26] | Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010). Light-regulated plant growth and development. Curr Top Dev Biol 91, 29-66. |
| [27] | Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999). Activation tagging of the floral inducer FT. Science 286, 1962-1965. |
| [28] | Krogan NT, Hogan K, Long JA (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180-4190. |
| [29] | Lau OS, Deng XW (2010). Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13, 571-577. |
| [30] | Li C, Qi LJ, Zhang SM, Dong XJ, Jing YJ, Cheng JK, Feng ZY, Peng J, Li H, Zhou YY, Wang XJ, Han R, Duan J, Terzaghi W, Lin RC, Li JG (2022). Mutual upregulation of HY5 and TZP in mediating phytochrome egsignaling. Plant Cell 34, 633-654. |
| [31] | Li JG, Hiltbrunner A (2021). Is the Pr form of phytochrome biologically active in the nucleus? Mol Plant 14, 535-537. |
| [32] | Li JG, Li G, Wang HY, Deng XW (2011). Phytochrome signaling mechanisms. Arabidopsis Book 2011, e0148. |
| [33] | Lin RC, Ding L, Casola C, Ripoll DR, Feschotte C, Wang HY (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318, 1302-1305. |
| [34] | Lin RC, Teng YB, Park HJ, Ding L, Black C, Fang P, Wang HY (2008). Discrete and essential roles of the multiple domains of Arabidopsis FHY3 in mediating phytochrome A signal transduction. Plant Physiol 148, 981-992. |
| [35] | Lopato S, Gattoni R, Fabini G, Stevenin J, Barta A (1999). A novel family of plant splicing factors with a Zn knuckle motif: examination of RNA binding and splicing activities. Plant Mol Biol 39, 761-773. |
| [36] | Loudet O, Michael TP, Burger BT, Le Mette C, Mockler TC, Weigel D, Chory J (2008). A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 105, 17193-17198. |
| [37] | Mlynárová L, Nap JP, Bisseling T (2007). The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J 51, 874-885. |
| [38] | Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011). The ELF4-ELF3- LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398-402. |
| [39] | Oh S, Zhang H, Ludwig P, Van Nocker S (2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16, 2940-2953. |
| [40] | Perrella G, Davidson MLH, O'Donnell L, Nastase AM, Herzyk P, Breton G, Pruneda-Paz JL, Kay SA, Chory J, Kaiserli E (2018). ZINC-FINGER interactions mediate transcriptional regulation of hypocotyl growth in Arabidopsis. Proc Natl Acad Sci USA 115, E4503-E4511. |
| [41] | Qu GP, Li H, Lin XL, Kong XX, Hu ZL, Jin YH, Liu Y, Song HL, Kim DH, Lin RC, Li JG, Jin JB (2020). Reversible SUMOylation of FHY1 regulates phytochrome A signaling in Arabidopsis. Mol Plant 13, 879-893. |
| [42] | Rizzini L, Favory JJ, Cloix C, Faggionato D, O'Hara A, Kaiserli E, Baumeister R, Sch?fer E, Nagy F, Jenkins GI, Ulm R (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103-106. |
| [43] | Saijo Y, Sullivan JA, Wang HY, Yang JP, Shen YP, Rubio V, Ma LG, Hoecker U, Deng XW (2003). The COP1- SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17, 2642-2647. |
| [44] | Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007). FKF1 and GIGANTEA complex formation is required for day- length measurement in Arabidopsis. Science 318, 261-265. |
| [45] | Szemenyei H, Hannon M, Long JA (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384-1386. |
| [46] | Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003-1006. |
| [47] | Vo LTA, Minet M, Schmitter JM, Lacroute F, Wyers F (2001). Mpe1, a zinc knuckle protein, is an essential component of yeast cleavage and polyadenylation factor required for the cleavage and polyadenylation of mRNA. Mol Cell Biol 21, 8346-8356. |
| [48] | Wang HY, Deng XW (2003). Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci 8, 172-178. |
| [49] | Wang L, Kim J, Somers DE (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 110, 761-766. |
| [50] | Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757-768. |
| [51] | Wier AD, Mayekar MK, Héroux A, Arndt KM, VanDemark AP (2013). Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc Natl Acad Sci USA 110, 17290-17295. |
| [52] | Xu DQ (2020). COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol 228, 1748-1753. |
| [53] | Yang CW, Xie FM, Jiang YP, Li Z, Huang X, Li L (2018). Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev Cell 44, 29-41. |
| [54] | Zhang BL, Wang L, Zeng LP, Zhang C, Ma H (2015). Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29, 975-987. |
| [55] | Zhang SM, Li C, Zhou YY, Wang XJ, Li H, Feng ZY, Chen HD, Qin GJ, Jin D, Terzaghi W, Gu HY, Qu LJ, Kang DM, Deng XW, Li JG (2018). TANDEM ZINC-FINGER/ PLUS3 is a key component of phytochrome A signaling. Plant Cell 30, 835-852. |
| [56] | Zhou YY, Yang L, Duan J, Cheng JK, Shen YP, Wang XJ, Han R, Li H, Li Z, Wang LH, Terzaghi W, Zhu DM, Chen HD, Deng XW, Li JG (2018). Hinge region of Arabidopsis phyA plays an important role in regulating phyA function. Proc Natl Acad Sci USA 115, E11864-E11873. |
/
| 〈 |
|
〉 |