

植物Aux/IAA基因家族生物学功能研究进展
收稿日期: 2021-09-24
录用日期: 2021-12-28
网络出版日期: 2021-12-28
基金资助
国家自然科学基金地区项目(32060451);浙江省自然科学基金重点项目(LZ19C020001)
Advances in Biological Functions of Aux/IAA Gene Family in Plants
Received date: 2021-09-24
Accepted date: 2021-12-28
Online published: 2021-12-28
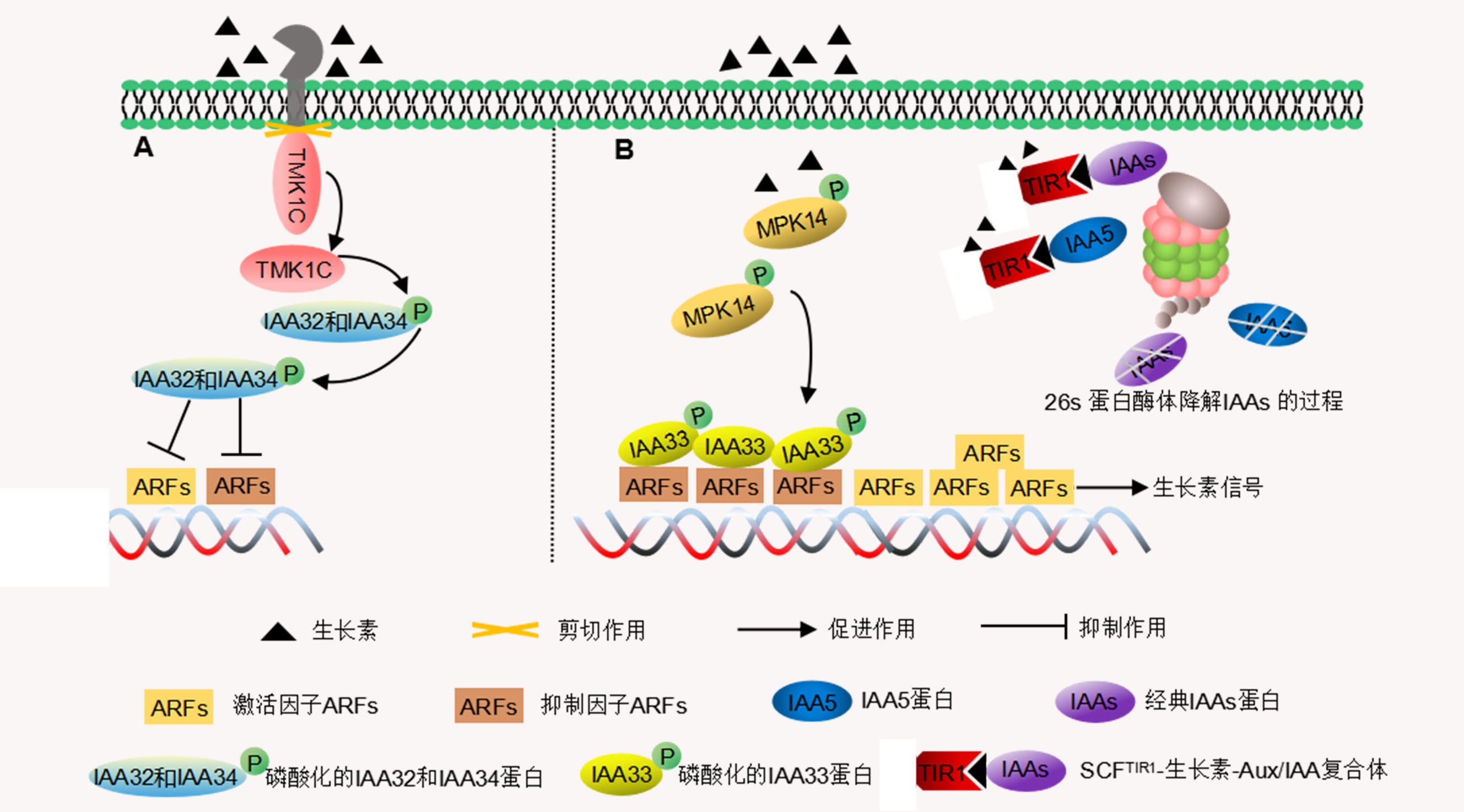
生长素是最重要的植物激素之一, 对植物生长发育起着关键调控作用。生长素作用于植物后, 早期生长素响应基因家族Aux/IAA、GH3和SAUR等被迅速诱导, 基因表达上调。其中Aux/IAA基因家族编码的蛋白一般由4个保守结构域组成, 结构域I具有抑制生长素信号下游基因表达的作用, 结构域II在生长素信号转导中主要被TIR1调控进而影响Aux/IAA的稳定性, 结构域III/IV通过与生长素响应因子ARF相互作用调控生长素信号。Aux/IAA基因家族在双子叶植物拟南芥(Arabidopsis thaliana)的器官发育、根形成、茎伸长和叶扩张等方面发挥重要作用; 在单子叶植物水稻(Oryza sativa)和小麦(Triticum aestivum)中, 主要影响根系发育和株型, 但大多数Aux/IAA基因的功能尚不清楚。该文主要从Aux/IAA蛋白的结构、功能和生长素信号转导途径方面综述Aux/IAA家族在拟南芥、禾谷类作物及其它植物中的研究进展, 以期为全面揭示Aux/IAA家族基因的生物学功能提供线索。
李艳艳 , 齐艳华 . 植物Aux/IAA基因家族生物学功能研究进展[J]. 植物学报, 2022 , 57(1) : 30 -41 . DOI: 10.11983/CBB21168
Auxin is one of the most important plant hormones and plays a key role in regulating plant growth and development. In plants, early auxin responsive gene families, such as Aux/IAA (Auxin/Indole acetic acid repressors), GH3 (Gretchen Hagen3) and SAUR (Small Auxin up RNA), are rapidly induced and up-regulated by auxin treatment. Aux/IAA gene family is generally composed of four conserved domains. Domain I inhibits the expression of downstream genes in the auxin signaling pathway, and domain II is mainly regulated by Transport Inhibitor Response 1 (TIR1) in auxin signal transduction, thus affecting the stability of Aux/IAA. Domain III/IV regulates auxin signaling by interacting with Auxin Response Factor (ARF). Aux/IAA gene family has been reported to play an important role in organ development, root formation, stem elongation and leaf expansion in dicotyledonous Arabidopsis thaliana while in monocotyledonous rice (Oryza sativa) and wheat (Triticum aestivum), Aux/IAA mainly affects root development and plant architecture. However, the functions of most Aux/IAA genes remain unclear and need to further study. In this article, we reviewed the structure and function of Aux/IAA protein, and the auxin signal transduction pathway in Arabidopsis, cereal crops and other plants to provide clues for fully revealing the biofunction of the Aux/IAA gene family.

Key words: Aux/IAA protein; auxin; Arabidopsis thaliana; biofunctions; cereal crops
| [1] | 韩悌倩 (2020). 干旱胁迫下马铃薯根系Aux/IAA转录因子的挖掘. 硕士论文. 兰州: 甘肃农业大学. pp. 3-4. |
| [2] | 胡晓, 侯旭, 袁雪, 管丹, 刘悦萍 (2017). ARF和Aux/IAA调控果实发育成熟机制研究进展. 生物技术通报 33(12), 37-44. |
| [3] | 林雨晴, 齐艳华 (2021). 生长素输出载体PIN家族研究进展. 植物学报 56, 151-165. |
| [4] | 刘艳娜 (2020). 小麦TaIAA8-1B基因的克隆与功能分析. 硕士论文. 青岛: 青岛大学. pp. 12-19. |
| [5] | 马军, 徐通达 (2020). 植物非经典生长素信号转导通路解析. 生物技术通报 36(7), 15-22. |
| [6] | 王婧 (2012). 拟南芥Aux/IAA家族基因IAA8参与花器官发育的研究. 博士论文. 武汉: 武汉大学. pp. 102-118. |
| [7] | 王闵霞 (2016). 水稻分蘖调控基因OsIAA16的功能研究和d14突变体遗传修饰因子的创制. 博士论文. 北京: 中国农业科学院. pp. 115-121. |
| [8] | 王曦烨 (2016). 棉花Aux/IAA家族基因功能的初步探究. 硕士论文. 河南: 河南师范大学. pp. 15-17. |
| [9] | 张战营 (2015). 水稻产量基因Gnp4的克隆与功能机理分析. 博士论文. 北京: 中国农业大学. pp. 97-109. |
| [10] | Abel S, Oeller PW, Theologis A (1994). Early auxin-indu-ced genes encode short-lived nuclear proteins. Proc Natl Acad Sci USA 91, 326-330. |
| [11] | Abel S, Theologis A (1995). A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J 8, 87-96. |
| [12] | Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M (2012). Genome-wide identification, functio-nal analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol 53, 659-672. |
| [13] | Bassa C, Mila I, Bouzayen M, Audran-Delalande C (2012). Phenotypes associated with down-regulation of Sl-IAA27support functional diversity among Aux/IAA family members in tomato. Plant Cell Physiol 53, 1583-1595. |
| [14] | Belin C, Megies C, Hauserová E, Lopez-Molina L (2009). Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21, 2253-2268. |
| [15] | Cao M, Chen R, Li P, Yu YQ, Zheng R, Ge DF, Zheng W, Wang XH, Gu YT, Gelová Z, Friml J, Zhang H, Liu RY, He J, Xu TD (2019). TMK1-mediated auxin signaling regulates differential growth of the apical hook. Nature 568, 240-243. |
| [16] | Chaaboun IS, Jones B, Delalande C, Wang H, Li ZG, Mila I, Frasse P, Latché A, Pech JC, Bouzayen M (2009). Sl- IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signaling involved in differential growth. J Exp Bot 60, 1349-1362. |
| [17] | Chen DD, Richardson T Chai SC, McIntyre CL, Rae AL, Xue GP (2016). Drought-up-regulated TaNAC69-1 is a transcriptional repressor of TaSHY2 and TaIAA7, and enhances root length and biomass in wheat. Plant Cell Physiol 57, 2076-2090. |
| [18] | Chen SH, Zhou LJ, Xu P, Xue HW (2018). SPOC domain- containing protein Leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling. PLoS Genet 14, e1007829. |
| [19] | Colón-Carmona A, Chen DL, Yeh KC, Abel S (2000). Aux/ IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol 124, 1728-1738. |
| [20] | Deng W, Yan F, Liu MC, Wang XY, Li ZG (2012). Down-regulation of SlIAA15 in tomato altered stem xylem development and production of volatile compounds in leaf exudates. Plant Signal Behav 7, 911-913. |
| [21] | Dharmasiri N, Dharmasiri S, Jones AM, Estelle M (2003). Auxin action in a cell-free system. Curr Biol 13, 1418-1422. |
| [22] | Friml J (2003). Auxin transport-shaping the plant. Curr Opin Plant Biol 6, 7-12. |
| [23] | Galli M, Liu QJ, Moss BL, Malcomber S, Li W, Gaines C, Federici S, Roshkovan J, Meeley R, Nemhauser JL, Gallavotti A (2015). Auxin signaling modules regulate maize inflorescence architecture. Proc Natl Acad Sci USA 112, 13372-13377. |
| [24] | Gao JP, Cao XL, Shi SD, Ma YL, Wang K, Liu SJ, Chen D, Chen Q, Ma HL (2016). Genome-wide survey of Aux/IAA gene family members in potato (Solanum tuberosum): identification, expression analysis, and evaluation of their roles in tuber development. Biochem Biophys Res Com-mun 471, 320-327. |
| [25] | Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H (2012). Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-media-ted auxin signaling. Philos Trans R Soc Lond B Biol Sci 367, 1461-1468. |
| [26] | Guilfoyle TJ (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27, 33-43. |
| [27] | Guilfoyle TJ, Hagen G (2007). Auxin response factors. Curr Opin Plant Biol 10, 453-460. |
| [28] | Hagen G, Guilfoyle T (2002). Auxin-responsive gene exp-ression: genes, promoters and regulatory factors. Plant Mol Biol 49, 373-385. |
| [29] | Han XY, Xu XY, Fang DD, Zhang TZ, Guo WZ (2012). Cloning and expression analysis of novel Aux/IAA family genes in Gossypium hirsutum. Gene 503, 83-91. |
| [30] | Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757-770. |
| [31] | Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001). The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchroni-a)zes pollen maturation, anther dehiscence, and flower ope-ning in Arabidopsis. Plant Cell 13, 2191-2209. |
| [32] | Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sati-va). Funct Integr Genomics 6, 47-59. |
| [33] | Jain M, Khurana JP (2009). Transcript profiling reveals diverse roles of auxin-responsive genes during reproduc-tive development and abiotic stress in rice. FEBS J 276, 3148-3162. |
| [34] | Jia ML, Li YN, Wang ZY, Tao S, Sun GL, Kong XC, Wang K, Ye XG, Liu SS, Geng SF, Mao L, Li AL (2021). TaIAA21 represses TaARF25-mediated expression of TaERFs required for grain size and weight development in wheat. Plant J 108, 1754-1767. |
| [35] | Jiang L, Li ZN, Yu XM, Liu CB (2021). Bioinformatics analysis of Aux/IAA gene family in maize. Agron J 113, 932-942. |
| [36] | John ME, Keller G (1996). Metabolic pathway engineering in cotton: biosynthesis of polyhydroxybutyrate in fiber cells. Proc Natl Acad Sci USA 93, 12768-12773. |
| [37] | Jung H, Lee DK, Choi YD, Kim JK (2015). OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci 236, 304-312. |
| [38] | Kitomi Y, Inahashi H, Takehisa H, Sato Y, Inukai Y (2012). OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Sci 190, 116-122. |
| [39] | Kloosterman B, Visser RGF, Bachem CWB (2006). Iso-lation and characterization of a novel potato Auxin/Indole-3- Acetic Acid family member (StIAA2) that is involved in petiole hyponasty and shoot morphogenesis. Plant Physiol Biochem 44, 766-775. |
| [40] | Leyser O (2018). Auxin signaling. Plant Physiol 176, 465-479. |
| [41] | Liscum E, Reed JW (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49, 387-400. |
| [42] | Liu JY, Shi MJ, Wang J, Zhang B, Li YS, Wang J, El- Sappah AH, Liang Y (2020). Comparative transcriptomic analysis of the development of sepal morphology in tomato (Solanum lycopersicum L.). Int J Mol Sci 21, 5914. |
| [43] | López-Bucio J, Ortiz-Castro R, Ruíz-Herrera LF, Juárez CV, Hernández-Madrigal F, Carreón-Abud Y, Martínez- Trujillo M (2015). Chromate induces adventitious root formation via auxin signaling and SOLITARY-ROOT/IAA-14 gene function in Arabidopsis thaliana. BioMetals 28, 353-365. |
| [44] | Lv BS, Yu QQ, Liu JJ, Wen XJ, Yan ZW, Hu KQ, Li HB, Kong XP, Li CL, Tian HY, De Smet I, Zhang XS, Ding ZJ (2020). Non-canonical AUX/IAA protein IAA33 compe-tes with canonical AUX/IAA repressor IAA5 to negatively regulate auxin signaling. EMBO J 39, e101515. |
| [45] | Mockaitis K, Estelle M (2008). Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24, 55-80. |
| [46] | Morgan KE, Zarembinski TI, Theologis A, Abel S (1999). Biochemical characterization of recombinant polypeptides corresponding to the predicted βααfold in Aux/IAA protein-s. FEBS Lett 454, 283-287. |
| [47] | Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, Ecker JR, Reed JW (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107-4118. |
| [48] | Nakamura A, Umemura I, Gomi K, Hasegawa Y, Kitano H, Sazuka T, Matsuoka M (2006). Production and characte-rization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J 46, 297-306. |
| [49] | Ni J, Wang GH, Zhu ZX, Zhang HH, Wu YR, Wu P (2011). OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J 68, 433-442. |
| [50] | Qiao JY, Jiang HZ, Lin YQ, Shang LG, Wang M, Li DM, Fu XD, Geisler M, Qi YH, Gao ZY, Qian Q (2021). A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol Plant 14, 1683-1698. |
| [51] | Qiao LY, Zhang XJ, Han X, Zhang L, Li X, Zhan HX, Ma J, Luo PG, Zhang WP, Cui L, Li XY, Chang ZJ (2015). A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.). Front Plant Sci 6, 770. |
| [52] | Reed JW (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6, 420-425. |
| [53] | Rogg LE, Lasswell J, Bartel B (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465-480. |
| [54] | Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371-1373. |
| [55] | Sakamoto T, Morinaka Y, Inukai Y, Kitano H, Fujioka S (2013). Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J 73, 676-688. |
| [56] | Salehin M, Li BH, Tang M, Katz E, Song L, Ecker JR, Kliebenstein DJ, Estelle M (2019). Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat Commun 10, 4021. |
| [57] | Singla B, Chugh A, Khurana JP, Khurana P (2006). An early auxin-responsive Aux/IAA gene from wheat (Triti-cum aestivum) is induced by epibrassinolide and differen-tially regulated by light and calcium. J Exp Bot 57, 4059-4070. |
| [58] | Smalle J, Vierstra RD (2004). The ubiquitin 26S proteaso-me proteolytic pathway. Annu Rev Plant Biol 55, 555-590. |
| [59] | Song YL, Xu ZF (2013). Ectopic overexpression of an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) gene OsIAA4 in rice induces morphological changes and reduces res-ponsiveness to auxin. Int J Mol Sci 14, 13645-13656. |
| [60] | Song YL, You J Xiong LZ (2009). Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol Biol 70, 297-309. |
| [61] | Su LY, Audran C, Bouzayen M, Roustan JP, Chervin C (2015). The Aux/IAA, Sl-IAA17 regulates quality parame-ters over tomato fruit development. Plant Signal Behav 10, e1071001. |
| [62] | Su LY, Bassa C, Audran C, Mila I, Cheniclet C, Chevalier C, Bouzayen M, Roustan JP, Chervin C (2014). The auxin Sl-IAA17 transcriptional repressor controls fruit size via the regulation of endoreduplication-related cell expan-sion. Plant Cell Physiol 55, 1969-1976. |
| [63] | Szemenyei H, Hannon M, Long JA (2008). TOPLESS mediates auxin-dependent transcriptional repression du-ring Arabidopsis embryogenesis. Science 319, 1384-1386. |
| [64] | Tan X, Calderon-Villalobos LIA, Sharon M, Zheng CX, Robinson CV, Estelle M, Zheng N (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640-645. |
| [65] | Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004). MASSU-GU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypo-cotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379-393. |
| [66] | Tian Q, Reed JW (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 ge-ne. Development 126, 711-721. |
| [67] | Timpte C, Wilson AK, Estelle M (1994). The axr2-1mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138, 1239-1249. |
| [68] | Tiwari SB, Hagen G, Guilfoyle TJ (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533-543. |
| [69] | Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001). AUX/ IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13, 2809-2822. |
| [70] | Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H (2008). Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Ara- bidopsis thaliana. Plant Cell Physiol 49, 1025-1038. |
| [71] | Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963-1971. |
| [72] | Von Behrens I, Komatsu M, Zhang YX, Berendzen KW, Niu XM, Sakai H, Taramino G, Hochholdinger F (2011). Rootless with undetectable meristem 1encodes a monocot-specific AUX/IAA protein that controls embryonic se-minal and post-embryonic lateral root initiation in maize. Plant J 66, 341-353. |
| [73] | Walker JC, Key JL (1982). Isolation of cloned cDNAs to auxin-responsive poly(A)+RNAs of elongating soybean hypocotyl. Proc Natl Acad Sci USA 79, 7185-7189. |
| [74] | Wang SK, Bai YH, Shen CJ, Wu YR, Zhang SN, Jiang DA, Guilfoyle TJ, Chen M, Qi YH (2010a). Auxin-related gene families in abiotic stress response in Sorghum bico-lor. Funct Integr Genomics 10, 533-546. |
| [75] | Wang YJ, Deng DX, Bian YL, Lv YP, Xie Q (2010b). Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays L.). Mol Biol Rep 37, 3991-4001. |
| [76] | Weijers D, Wagner D (2016). Transcriptional responses to the auxin hormone. Annu Rev Plant Biol 67, 539-574. |
| [77] | Wilson AK, Pickett FB, Turner JC, Estelle M (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222, 377-383. |
| [78] | Woodward AW, Bartel B (2005). Auxin: regulation, action, and interaction. Ann Bot 95, 707-735. |
| [79] | Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theo- logis A, Callis J (2000). Degradation of Aux/IAA proteins is essential for normal auxin signaling. Plant J 21, 553-562. |
| [80] | Yang XQ, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M (2004). The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J 40, 772-782. |
| [81] | Zhang AY, Yang X, Lu J, Song FY, Sun JH, Wang C, Lian J, Zhao LL, Zhao BC (2021). OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci 308, 110903. |
| [82] | Zhang JH, Chen RG, Xiao JH, Qian CJ, Wang TT, Li HX, Ouyang B Ye ZB (2007). A single-base deletion mutation in SlIAA9 gene causes tomato (Solanum lycopersicum) entire mutant. J Plant Res 120, 671-678. |
| [83] | Zhang SN, Wang SK, Xu YX, Yu CL, Shen CJ, Qian Q, Geisler M, Jiang DA, Qi YH (2015). The auxin response factor, OsARF19, controls rice leaf angles through positi-vely regulating OsGH3-5 and OsBRI1. Plant Cell Environ 38, 638-654. |
| [84] | Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, Lohmann JU (2010). Hormonal control of the shoot stem-cell niche. Nature 465, 1089-1092. |
| [85] | Zhu ZX, Liu Y, Liu SJ, Mao CZ, Wu YR, Wu P (2012). A gain-of-function mutation in OsIAA11affects lateral root development in rice. Mol Plant 5, 154-161. |
/
| 〈 |
|
〉 |