

植物小RNA荧光原位杂交实验方法
收稿日期: 2020-04-02
录用日期: 2021-05-07
网络出版日期: 2021-05-07
基金资助
中国科学院战略性先导科技专项(A类) No(XDA24010106-2);国家自然科学基金No(31900391)
Protocols for Small RNA FISH in Plants
Received date: 2020-04-02
Accepted date: 2021-05-07
Online published: 2021-05-07
胡滨滨 , 薛治慧 , 张翠 . 植物小RNA荧光原位杂交实验方法[J]. 植物学报, 2021 , 56(3) : 330 -338 . DOI: 10.11983/CBB21057
Small RNAs are a type of small nucleotide molecules that are essential for plant growth and development, playing a key role in variety of life processes and in response to stresses. Research on the location of small RNAs could discover their functions in plants. Small RNA FISH is a qualitative or semi-quantitative analysis of small RNA in organisms by fluorescence detection technology. At present, this technology has been widely used in animals, but it is still less applied in plants. This article introduces the specific operation procedures and attentions based on ultra-high resolution microscopy that combines locked nucleic acid (LNA) probe in situ hybridization with immunofluorescence. This protocol can be used to detect the expression and localization of small RNA in plant tissues.
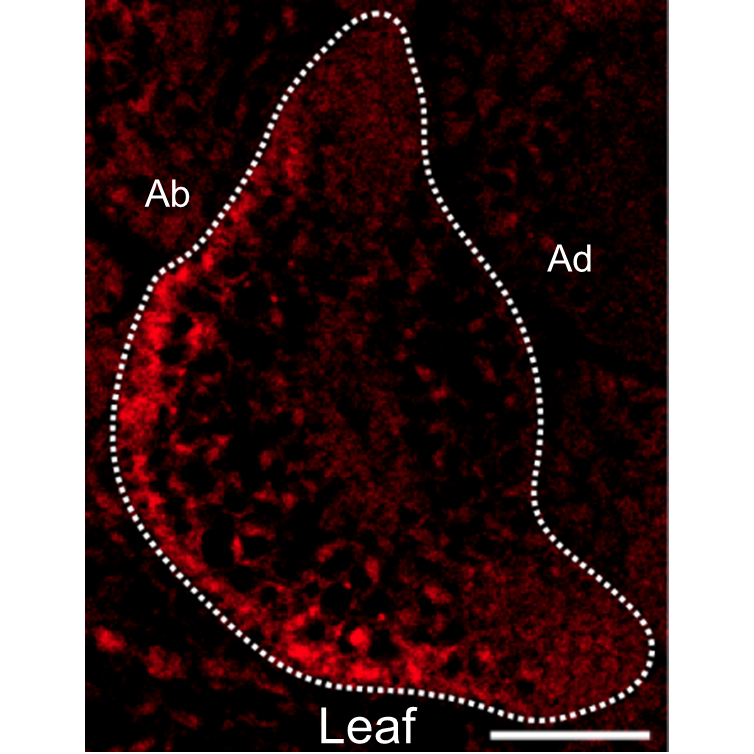
Key words: fluorescent in situ hybridization; small RNA; plants
| 1 | Baumberger N, Baulcombe DC (2005). Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102, 11928-11933. |
| 2 | Bhogale S, Mahajan AS, Natarajan B, Rajabhoj M, Thulasiram HV, Banerjee AK (2014). MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol 164, 1011-1027. |
| 3 | Bleckmann A, Dresselhaus T (2016). Fluorescent whole- mount RNA in situ hybridization (F-WISH) in plant germ cells and the fertilized ovule. Methods 98, 66-73. |
| 4 | Bruno L, Muto A, Spadafora ND, Iaria D, Chiappetta A, Van Lijsebettens M, Bitonti MB (2011). Multi-probe in situ hybridization to whole mount Arabidopsis seedlings. Int J Dev Biol 55, 197-203. |
| 5 | Bruno L, Ronchini M, Gagliardi O, Corinti T, Chiappetta A, Gerola P, Bitonti MB (2015). Analysis of ATGUS1 and ATGUS2 in Arabidopsis root apex by a highly sensitive TSA-MISH method. Int J Dev Biol 59, 221-228. |
| 6 | Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J (2008). Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J 53, 739-749. |
| 7 | Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, Campilho A, Sebastian J, Bowman JL, Helariutta Y, Benfey PN (2010). Cell signaling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316-321. |
| 8 | Chen XM (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022-2025. |
| 9 | Chen XM (2009). Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25, 21-44. |
| 10 | Chitwood DH, Nogueira FT, Howell MD, Montogmery TA, Carrington JC, Timmermans MC (2008). Pattern formation in leaves via small RNA mobility. Dev Biol 319, 587-588. |
| 11 | Chitwood DH, Nogueira FTS, Howell MD, Montgomery TA, Carrington JC, Timmermans MCP (2009). Pattern formation via small RNA mobility. Gene Dev 23, 549-554. |
| 12 | Fischer AH, Jacobson KA, Rose J, Zeller R (2008). Paraffin embedding tissue samples for sectioning. CSH Protoc 2008, pdb.prot4989. |
| 13 | Himanen K, Woloszynska M, Boccardi TM, De Groeve S, Nelissen H, Bruno L, Vuylsteke M, Van Lijsebettens M (2012). Histone H2b monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J 72, 249-260. |
| 14 | Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidop- sis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19, 926-942. |
| 15 | Huang K, Baldrich P, Meyers BC, Caplan JL (2019). sRNA-FISH: versatile fluorescent in situ detection of small RNAs in plants. Plant J 98, 359-369. |
| 16 | Huen AK, Rodriguez-Medina C, Ho AYY, Atkins CA, Smith PMC (2017). Long-distance movement of phospha- te starvation-responsive microRNAs in Arabidopsis. Plant Biol 19, 643-649. |
| 17 | Javelle M, Timmermans MCP (2012). In situ localization of small RNAs in plants by using LNA probes. Nat Protoc 7, 533-541. |
| 18 | Jiang JM (2019). Fluorescence in situ hybridization in plants: recent developments and future applications. Chromoso- me Res 27, 153-165. |
| 19 | Jones-Rhoades MW, Bartel DP, Bartel B (2006). Micro- RNAs and their regulatory roles in plants. Annu Rev Plant Biol 57, 19-53. |
| 20 | Jung JH, Park CM (2007). MIR166/ 165 genes exhibit dynamic expression patterns in regulating shoot apical meris- tem and floral development in Arabidopsis. Planta 225, 1327-1338. |
| 21 | Kidner C, Timmermans M (2006). In situ hybridization as a tool to study the role of microRNAs in plant development. Methods Mol Biol 342, 159-179. |
| 22 | Kim VN, Han JJ, Siomi MC (2009). Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10, 126-139. |
| 23 | Knauer S, Holt AL, Rubio-Somoza I, Tucker EJ, Hinze A, Pisch M, Javelle M, Timmermans MC, Tucker MR, Laux T (2013). A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev Cell 24, 125-132. |
| 24 | Kurihara Y, Watanabe Y (2004). Arabidopsis micro-RNA biogenesis through dicer-like 1 protein functions. Proc Natl Acad Sci USA 101, 12753-12758. |
| 25 | Lamb JC, Danilova T, Bauer MJ, Meyer JM, Holland JJ, Jensen MD, Birchler JA (2007). Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics 175, 1047-1058. |
| 26 | Lee Y, Kim M, Han JJ, Yeom KH, Lee S, Baek SH, Kim VN (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23, 4051-4060. |
| 27 | Levsky JM, Singer RH (2003). Fluorescence in situ hybridization: past, present and future. J Cell Sci 116, 2833-2838. |
| 28 | Li JJ, Yang ZY, Yu B, Liu J, Chen XM (2005). Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis. Curr Biol 15, 1501-1507. |
| 29 | Li S, Wang XT, Xu WY, Liu T, Cai CM, Chen LY, Clark CB, Ma JX (2021). Unidirectional movement of small RNAs from shoots to roots in interspecific heterografts. Nat Plants 7, 50-59. |
| 30 | Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147, 732-746. |
| 31 | Liu QL, Yao XZ, Pi LM, Wang H, Cui XF, Huang H (2009). The ARGONAUTE10 gene modulates shoot apical meris- tem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J 58, 27-40. |
| 32 | Lu J, Tsourkas A (2009). Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res 37, e100. |
| 33 | Moissiard G, Parizotto EA, Himber C, Voinnet O (2007). Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 13, 1268-1278. |
| 34 | Nodine MD, Bartel DP (2010). MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Gene Dev 24, 2678-2692. |
| 35 | Nogueira FTS, Chitwood DH, Madi S, Ohtsu K, Schnable PS, Scanlon MJ, Timmermans MCP (2009). Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet 5, e1000320. |
| 36 | Nogueira FTS, Madi S, Chitwood DH, Juarez MT, Timmer- mans MCP (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Gene Dev 21, 750-755. |
| 37 | Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, Alvarez JP, Blum E, Zamir D, Eshed Y (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39, 787-791. |
| 38 | Pagliarani C, Gambino G (2019). Small RNA mobility: spread of RNA silencing effectors and its effect on developmental processes and stress adaptation in plants. Int J Mol Sci 20, 4306. |
| 39 | Pant BD, Buhtz A, Kehr J, Scheible WR (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53, 731-738. |
| 40 | Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR (2009). Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150, 1541-1555. |
| 41 | Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Gene Dev 18, 2237-2242. |
| 42 | Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS (2005). Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA 102, 3691-3696. |
| 43 | Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55, 65-76. |
| 44 | Rozier F, Mirabet V, Vernoux T, Das P (2014). Analysis of 3D gene expression patterns in plants using whole-mount RNA in situ hybridization. Nat Protoc 9, 2464-2475. |
| 45 | Skopelitis DS, Hill K, Klesen S, Marco CF, von Born P, Chitwood DH, Timmermans MCP (2018). Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nat Commun 9, 3107. |
| 46 | Tirichine L, Andrey P, Biot E, Maurin Y, Gaudin V (2009). 3D fluorescent in situ hybridization using Arabidopsis leaf cryosections and isolated nuclei. Plant Methods 5, 11. |
| 47 | Vaucheret H, Vazquez F, Crété P, Bartel DP (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Gene Dev 18, 1187-1197. |
| 48 | Wollmann H, Mica E, Todesco M, Long JA, Weigel D (2010). On reconciling the interactions between APETA- LA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633-3642. |
| 49 | Woloszynska M, Le Gall S, Neyt P, Boccardi TM, Grasser M, L?ngst G, Aesaert S, Coussens G, Dhondt S, Van De Slijke E, Bruno L, Fung-Uceda J, Mas P, Van Montagu M, Inzé D, Himanen K, De Jaeger G, Grasser KD, Van Lijsebettens M (2019). Histone 2B monoubiquitination complex integrates transcript elongation with RNA processing at circadian clock and flowering regulators. Proc Natl Acad Sci USA 116, 8060-8069. |
| 50 | Xue ZH, Liu LY, Zhang C (2020). Regulation of shoot apical meristem and axillary meristem development in plants. Int J Mol Sci 21, 2917. |
| 51 | Yang WB, Schuster C, Prunet N, Dong QK, Landrein B, Wightman R, Meyerowitz EM (2020). Visualization of protein coding, long noncoding, and nuclear RNAs by fluorescence in situ hybridization in sections of shoot apical meristems and developing flowers. Plant Physiol 182, 147-158. |
| 52 | Yang WB, Wightman R, Meyerowitz EM (2017). Cell cycle control by nuclear sequestration of CDC20 and CDH1 mRNA in plant stem cells. Mol Cell 68, 1108-1119. |
| 53 | Yu Y, Zhang YC, Chen XM, Chen YQ (2019). Plant noncoding RNAs: hidden players in development and stress responses. Annu Rev Cell Dev Biol 35, 407-431. |
| 54 | Zhang C, Fan LS, Le BH, Ye PY, Mo BX, Chen XM (2020). Regulation of ARGONAUTE10 expression enables temporal and spatial precision in axillary meristem initiation in Arabidopsis. Dev Cell 55, 603-616. |
| 55 | Zhang C, Wang J, Wenkel S, Chandler JW, Werr W, Jiao YL (2018). Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 145, dev158352. |
/
| 〈 |
|
〉 |