

基于双碱基编辑系统的植物基因靶向随机突变技术
收稿日期: 2021-01-11
录用日期: 2021-02-25
网络出版日期: 2021-02-25
基金资助
中国科学院战略性先导专项(XDA24020102);国家自然科学基金(31788103);国家自然科学基金(31900301);国家重点研发计划(2016 YFD0101804)
Saturation Mutagenesis Using Dual Cytosine and Adenine Base Editors
Received date: 2021-01-11
Accepted date: 2021-02-25
Online published: 2021-02-25
张瑞 , 高彩霞 . 基于双碱基编辑系统的植物基因靶向随机突变技术[J]. 植物学报, 2021 , 56(1) : 50 -55 . DOI: 10.11983/CBB21009
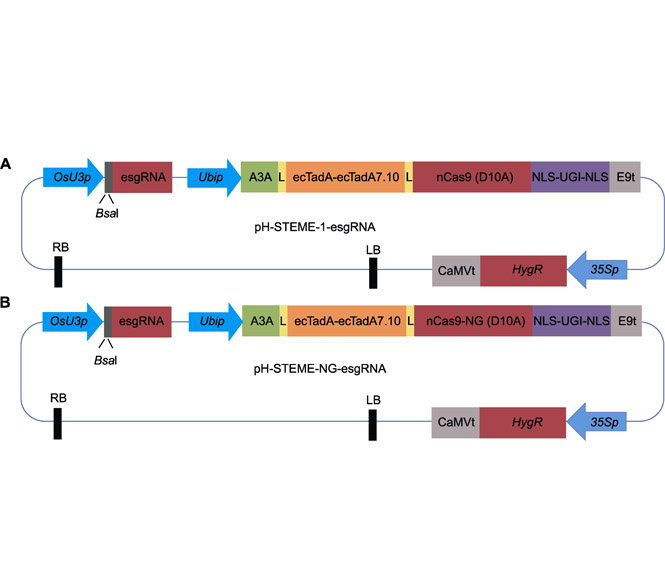
Because the genome of an organism determines its primary phenotype, evolutionary principles suggest that genetic variations enhance phenotypic diversity towards increased fitness. Targeted saturation mutagenesis of crop genes could be used to screen for genetic variants with improved agronomic traits. Compared to traditional mutational breeding or directed evolution in heterologous organisms, targeted mutagenesis via dual cytosine and adenine base editors effectively generates endogenous mutagenesis and facilitates in vivo directed evolution of plant genes. In this protocol, we detail the process towards using saturated targeted endogenous mutagenesis editors (STEMEs) to generate targeted, random mutagenesis of plant genes. In particular, we focus on the process of designing targets, screening and genotyping the resulting evolved variants.
| [1] | Engqvist MKM, Rabe KS (2019). Applications of protein engineering and directed evolution in plant research. Plant Physiol 179, 907-917. |
| [2] | Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017). Programmable base editing of A?T to G?C in genomic DNA without DNA cleavage. Nature 551, 464-471. |
| [3] | Gehrke JM, Cervantes O, Clement MK, Wu YX, Zeng J, Bauer DE, Pinello L, Joung JK (2018). An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat Biotechnol 36, 977-982. |
| [4] | Henikoff S, Till BJ, Comai L (2004). TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol 135, 630-636. |
| [5] | Li C, Zhang R, Meng XB, Chen S, Zong Y, Lu CJ, Qiu JL, Chen YH, Li JY, Gao CX (2020). Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat Biotechnol 38, 875-882. |
| [6] | Li C, Zong Y, Wang YP, Jin S, Zhang DB, Song QN, Zhang R, Gao CX (2018). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19, 59. |
| [7] | Liu Q, Wang C, Jiao XZ, Zhang HW, Song LL, Li YX, Gao CX, Wang KJ (2019). Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cassystems. Sci China Life Sci 62, 1-7. |
| [8] | Meng XB, Yu H, Zhang Y, Zhuang FF, Song XG, Gao SS, Gao CX, Li JY (2017). Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol Plant 10, 1238-1241. |
| [9] | Nishimasu H, Shi X, Ishiguro S, Gao LY, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O (2018). Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259-1262. |
| [10] | Pacher M, Puchta H (2017). From classical mutagenesis to nuclease-based breeding-directing natural DNA repair for a natural end-product. Plant J 90, 819-833. |
| [11] | Packer MS, Liu DR (2015). Methods for the directed evolution of proteins. Nat Rev Genet 16, 379-394. |
| [12] | Shan QW, Wang YP, Chen KL, Liang Z, Li J, Zhang Y, Zhang K, Liu JX, Voytas DF, Zheng XL, Zhang Y, Gao CX (2013). Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol Plant 6, 1365-1368. |
| [13] | Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D (2005). A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat Biotechnol 23, 75-81. |
| [14] | Zong Y, Song QN, Li C, Jin S, Zhang DB, Wang YP, Qiu JL, Gao CX (2018). Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat Biotechnol 36, 950-953. |
/
| 〈 |
|
〉 |