

芒果胶孢炭疽病菌应答菌丝机械损伤产生无性孢子的分子机制
收稿日期: 2020-02-22
录用日期: 2020-05-08
网络出版日期: 2020-05-12
基金资助
海南省科技厅创新团队(2019CXTD399)
Molecular Mechanism of the Generation of Asexual Spores of the Mango Fungal Pathogen (Colletotrichum gloeosporioides) Induced by Mechanical Injuries
Received date: 2020-02-22
Accepted date: 2020-05-08
Online published: 2020-05-12
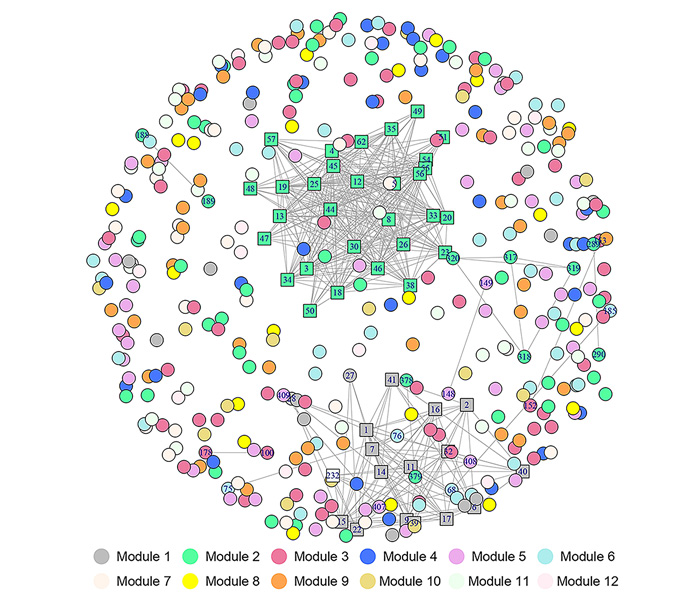
胶孢炭疽菌(Colletotrichum gloeosporioides)是引发芒果(Mangifera indica)炭疽病的主要病原体。室内平板培养胶孢炭疽菌不产生或产生很少分生孢子的情况时有发生, 但菌丝在机械损伤后24-48小时会产生大量分生孢子。胶孢炭疽菌应答机械损伤诱导产孢的核心基因及关键代谢通路尚未见报道。基于转录组测序(RNA-seq)技术检测了芒果胶孢炭疽菌菌丝在机械损伤处理后2小时内5个时间点的基因表达变化, 对差异表达基因进行GO富集和KEGG代谢通路富集分析, 并对菌丝响应胁迫的基因动态表达数据进行分析。基于常微分方程ODE模型结合变量选择技术, 构建了动态基因调控网络。结果表明, 有417个差异表达基因参与应答胶孢炭疽菌菌丝机械损伤, 分属12个聚类模块, 有4条通路存在显著富集, 分别是丙酮酸代谢、硫代谢、黄曲霉素合成途径和二萜合成途径。结合功能注释筛选出12个应答菌丝损伤胁迫的核心基因。研究结果为后续深入开展芒果胶孢炭疽菌产孢和致病机理研究奠定了重要基础。
王丽妍, 卢梦瑶, 童悦, 徐祥斌, 张正科, 孟兰环, 史学群, 宋海超 . 芒果胶孢炭疽病菌应答菌丝机械损伤产生无性孢子的分子机制[J]. 植物学报, 2020 , 55(5) : 551 -563 . DOI: 10.11983/CBB20026
Colletotrichum gloeosporioides is a prevalent pathogen that causes anthracnose in mango (Mangifera indica). Mycelium of C. gloeosporioides will accumulate a large number of conidia in 24-48 hours after mechanical injuries. However, it often accumulates none or few conidia during indoor culture, and the gene regulatory networks of the response to injury for a short-time (ST), or the key metabolic pathways involved in the response has not been explored. In this study, RNA-seq was carried out on RNA samples obtained at 5 time points within 2 hours after mechanical injuries. The differentially expressed genes were enriched by GO enrichment and KEGG metabolic pathway. The expression dynamics of mycelia in response to ST injury stress was analyzed. Based on a nonlinear ordinary differential equation model coupled with variable selection techniques, inter-module networks were constructed. The results showed that 417 differentially expressed genes were obtained, which belong to 12 clustered modules. KEGG enrichment analysis of differentially expressed genes was enriched in the process of pyruvate metabolism, sulfur metabolism, aflatoxin biosynthesis, diterpenoid biosynthesis. Combined with functional annotation, 12 core genes were identified that significantly correlated with ST injury-induced expression. These results provide valuable references for further research on asexual development and pathogenicity in C. gloeosporioides.

| [1] | 范玲玲, 陈刚, 陈义芳, 周卫东, 戴绍军, 孙国荣 ( 2010). NaHCO3胁迫下星星草根中Ca 2+与Ca 2+-ATPase的超微细胞化学定位 . 植物学报 45, 337-344. |
| [2] | 黄华平 ( 2016). 柱花草胶孢炭疽菌基因组、转录组测序及致病相关基因CgALS分析研究. 博士论文. 海口: 海南大学. pp. 1-115. |
| [3] | 李伟 ( 2004). 芒果胶孢炭疽病菌遗传转化体系的建立. 硕士论文. 海口: 华南热带农业大学. pp. 1-55. |
| [4] | 吴振麟 ( 2011). 芒果采后生理及贮藏保鲜技术研究进展. 安徽农学通报 17(19), 82-84, 135. |
| [5] | 于海英, 兰建强, 王晓燕, 彭磊, 吴佳, 刘云龙 ( 2012). 芒果胶孢炭疽菌致病性的初步研究. 见: 中国植物病理学会2012年学术年会论文集. 青岛: 中国植物病理学会. pp. 186-189. |
| [6] | 张丽勍, 段可, 邹小花, 何成勇, 高清华 ( 2017). 草莓胶孢炭疽菌CFEM候选效应子的生物信息学鉴定及其侵染过程中的转录分析. 植物保护 43(5), 43-51. |
| [7] | 章树桃, 陈春, 解廷娜 ( 2017). 新蚜虫疠霉菌丝和孢子阶段的转录组学分析. 农业生物技术学报 25, 1489-1499. |
| [8] | 周韬 ( 2014). 盐胁迫下胡杨的生理响应及miRNA表达动态变化. 博士论文. 北京: 北京林业大学. pp. 1-138. |
| [9] | 邹日娥 ( 1995). 芒果采后生理生化的变化. 东南园艺 ( 2), 9-12. |
| [10] | Adams TH, Boylan MT, Timberlake WE ( 1988). brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54, 353-362. |
| [11] | Bhetariya PJ, Prajapati M, Bhaduri A, Mandal RS, Varma A, Madan T, Singh Y, Sarma PU ( 2016). Phylogenetic and structural analysis of polyketide synthases in Aspergilli. Evol Bioinform 12, 109-119. |
| [12] | Chang C, Wu P, Baker RE, Maini PK, Alibardi L, Chuong CM ( 2009). Reptile scale paradigm: evo-devo, pattern formation and regeneration. Int J Dev Biol 53, 813-826. |
| [13] | Chi MH, Park SY, Kim S, Lee YH ( 2009). A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog 5, e1000401. |
| [14] | Conesa A, G?tz S, García-Gómez JM, Terol J, Talón M, Robles M ( 2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674-3676. |
| [15] | Elela SA, Nazar RN ( 1997). Role of the 5.8S rRNA in ribosome translocation. Nucleic Acids Res 25, 1788-1794. |
| [16] | Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, González VM, Hénaff E, Camara F, Cozzuto L, Lowy E, Alioto T, Capella-Gutiérrez S, Blanca J, Ca?izares J, Ziarsolo P, Gonzalez-Ibeas D, Rodríguez- Moreno L, Droege M, Du L, Alvarez-Tejado M, Lorente-Galdos B, Melé M, Yang LM, Weng YQ, Navarro A, Marques-Bonet T, Aranda MA, Nuez F, Picó B, Gabaldón T, Roma G, Guigó R, Casacuberta JM, Arús P, Puigdomènech P ( 2012). The genome of melon (Cucumis melo L.). Proc Natl Acad Sci USA 109, 11872-11877. |
| [17] | Hernandez CEM, Guerrero IEP, Hernandez GAG, Solis ES, Guzman JCT ( 2010). Catalase overexpression reduces the germination time and increases the pathogenicity of the fungus Metarhizium anisopliae. Appl Microbiol Biotechnol 87, 1033-1044. |
| [18] | Hernández-O?ate MA, Esquivel-Naranjo EU, Mendoza- Mendoza A, Stewart A, Herrera-Estrella AH ( 2012). An injury-response mechanism conserved across kingdoms determines entry of the fungus Trichoderma atroviride into development. Proc Natl Acad Sci USA 109, 14918-14923. |
| [19] | Jahurul MHA, Zaidul ISM, Ghafoor K, Al-Juhaimi FY, Nyam KL, Norulaini NAN, Sahena F, Mohd Omar AK ( 2015). Mango ( Mangifera indica L.) by-products and their valuable components: a review. Food Chem 183, 173-180. |
| [20] | Jedd G ( 2011). Fungal evo-devo: organelles and multicellular complexity. Trends Cell Biol 21, 12-19. |
| [21] | Jin JP, Zhang H, Kong L, Gao G, Luo JC ( 2014). PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42, D1182-D1187. |
| [22] | Kavanagh KL, J?rnvall H, Persson B, Oppermann U ( 2008). Medium-and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci 65, 3895-3906. |
| [23] | Langmead B, Salzberg SL ( 2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357-359. |
| [24] | Livak KJ, Schmittgen TD ( 2001). Analysis of relative gene expression data using real-time quantitative PCR and the ${{2}^{-\Delta \Delta \text{CT}}}$ method . Methods 25, 402-408. |
| [25] | McCarthy DJ, Chen YS, Smyth GK ( 2012). Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288-4297. |
| [26] | Munns R (2010). Approaches to identifying genes for salinity tolerance and the importance of timescale. In: Sunkar R, ed. Plant Stress Tolerance. New York: Humana Press. pp. 25-38. |
| [27] | Persson B, Hedlund J, J?rnvall H ( 2008). Medium-and short-chain dehydrogenase/reductase gene and protein families: the MDR superfamily. Cell Mol Life Sci 65, 3879-3894. |
| [28] | Perumal AB, Sellamuthu PS, Nambiar RB, Sadiku ER ( 2017). Effects of essential oil vapour treatment on the postharvest disease control and different defence responses in two mango ( Mangifera indica L.) cultivars. Food Bioprocess Technol 10, 1131-1141. |
| [29] | Qiu Q, Ma T, Hu QJ, Liu BB, Wu YX, Zhou HH, Wang Q, Wang J, Liu JQ ( 2011). Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol 31, 452-461. |
| [30] | Rendic S, Guengerich FP ( 2015). Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem Res Toxicol 28, 38-42. |
| [31] | Reyes-Perez JJ, Hernandez-Montiel LG, Vero S, Noa-Carrazana JC, Qui?ones-Aguilar EE, Rincón-Enríquez G ( 2019). Postharvest biocontrol of Colletotrichum gloeosporioides on mango using the marine bacterium Stenotrophomonas rhizophila and its possible mechanisms of action. J Food Sci Technol 56, 4992-4999. |
| [32] | Robinson MD, McCarthy DJ, Smyth GK ( 2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. |
| [33] | Saier MH Jr, Reddy VS, Tamang DG, V?stermark ? ( 2014). The transporter classification database. Nucleic Acids Res 42, D251-D258. |
| [34] | Sakamoto H, Matsuda O, Iba K ( 2008). ITN1, a novel gene encoding an ankyrin-repeat protein that affects the ABA-mediated production of reactive oxygen species and is involved in salt-stress tolerance in Arabidopsis thaliana. Plant J 56, 411-422. |
| [35] | Shao YZ, Zeng JK, Tang H, Zhou Y, Li W ( 2019). The chemical treatments combined with antagonistic yeast control anthracnose and maintain the quality of postharvest mango fruit. J Integr Agric 18, 1159-1169. |
| [36] | Shimizu Y, Ogata H, Goto S ( 2017). Type III polyketide synthases: functional classification and phylogenomics. Chembiochem 18, 50-65. |
| [37] | Si YQ, Liu P, Li PH, Brutnell TP ( 2014). Model-based clustering for RNA-seq data. Bioinformatics 30, 197-205. |
| [38] | Sun J, Chen SL, Dai SX, Wang RG, Li NY, Shen X, Zhou XY, Lu CF, Zheng XJ, Hu ZM, Zhang ZK, Song J, Xu Y ( 2009). NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiol 149, 1141-1153. |
| [39] | The Gene Ontology Consortium ( 2015). Gene ontology consortium: going forward. Nucleic Acids Res 43, D1049-D1056. |
| [40] | Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L ( 2010). Transcript assembly and quantification by RNA- seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511-515. |
| [41] | Wakabayashi K, Soga K, Hoson T ( 2012). Phenylalanine ammonia-lyase and cell wall peroxidase are cooperatively involved in the extensive formation of ferulate network in cell walls of developing rice shoots. J Plant Physiol 169, 262-267. |
| [42] | Wilczynski B, Furlong EEM ( 2010). Challenges for modeling global gene regulatory networks during development: insights from Drosophila. Dev Biol 340, 161-169. |
| [43] | Xie C, Mao XZ, Huang JJ, Ding Y, Wu JM, Dong S, Kong L, Gao G, Li CY, Wei LP ( 2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39, W316-W322. |
| [44] | Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA ( 2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508-522. |
| [45] | Yan JQ, Wang J, Zhang H ( 2002). An ankyrin repeat-containing protein plays a role in both disease resistance and antioxidation metabolism. Plant J 29, 193-202. |
| [46] | Zeng FR, Wu XJ, Qiu BY, Wu FB, Jiang LX, Zhang GP ( 2014). Physiological and proteomic alterations in rice ( Oryza sativa L.) seedlings under hexavalent chromium stress. Planta 240, 291-308. |
/
| 〈 |
|
〉 |