miR172-AP2模块调控植物生长发育及逆境响应的研究进展
- 扬州大学农学院, 江苏省作物基因组学和分子育种重点实验室/植物功能基因组学教育部重点实验室/粮食作物现代产业技术协同创新中心, 扬州 225009
收稿日期: 2019-09-11
录用日期: 2019-12-19
网络出版日期: 2019-12-19
基金资助
国家转基因重大科技专项(2016ZX08009003-004);江苏省高等学校自然科学研究重大项目(No.17KJA210001);江苏省六大人才高峰(No.SWYY-154)
Advances in the Regulation of Plant Growth and Development and Stress Response by miR172-AP2 Module
- Co-innovation Center for Modern Production Technology of Grain Crops/Key Laboratory of Plant Functional Genomics of the Ministry of Education/Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding, College of Agriculture, Yangzhou University, Yangzhou 225009, China
Received date: 2019-09-11
Accepted date: 2019-12-19
Online published: 2019-12-19
摘要
本文引用格式
王劲东 , 周豫 , 余佳雯 , 范晓磊 , 张昌泉 , 李钱峰 , 刘巧泉 . miR172-AP2模块调控植物生长发育及逆境响应的研究进展[J]. 植物学报, 2020 , 55(2) : 205 -215 . DOI: 10.11983/CBB19177
Abstract
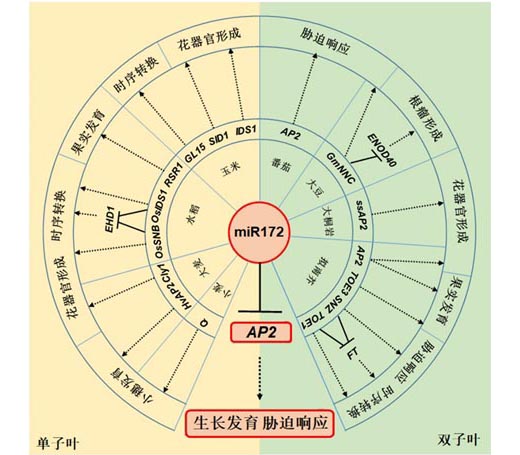
MicroRNA (miRNA), a kind of regulatory non-coding small RNA, induces degradation of target mRNA or inhibits its translation by specific or non-specific binding, thereby regulating plant growth and development. AP2, the target of miR172, encodes transcription factors that are unique to plants. miR172 regulates the expression of AP2 at the post-transcriptional or translational levels, thus regulating plant floral development, phase transition, spikelet morphology, tuber and fruit development, nodulation in legumes and stress response. Here we summarize the recent advances in the regulation of plant growth and development by miR172-AP2 regulatory module.
参考文献
| [1] | 陈丽, 鲁海琴, 李日慧, 傅廷栋, 沈金雄 ( 2018). 油菜miRNA研究现状与展望. 中国油料作物学报 40, 664-673. |
| [2] | 董淼, 黄越, 陈文铎, 徐涛, 郎秋蕾 ( 2013). 降解组测序技术在植物miRNA研究中的应用. 植物学报 48, 344-353. |
| [3] | 刘炜婳, 林玉玲, 林争春, 倪珊珊, 赖钟雄 ( 2018). 植物miR172家族成员进化与分子特性分析. 热带作物学报 39, 525-533. |
| [4] | 王幼宁, 苏超, 邹艳敏, 王利祥, 李霞 ( 2016). microRNA172参与植物生长发育及逆境响应的研究进展. 生命科学 28, 645-654. |
| [5] | 张俊红, 张守攻, 齐力旺, 童再康 ( 2014). 植物成熟microRNA转录后修饰与降解的研究进展. 植物学报 49, 483-489. |
| [6] | 赵晓晖, 孔凡江, 刘宝辉 ( 2017). 植物miR172及其靶基因调控开花与发育的研究进展. 黑龙江农业科学 ( 2), 126-130. |
| [7] | Acosta IF, Laparra H, Romero SP, Schmelz E, Hamberg M, Mottinger JP, Moreno MA, Dellaporta SL ( 2009). tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323, 262-265. |
| [8] | Agarwal P, Agarwal PK, Joshi AJ, Sopory SK, Reddy MK ( 2010). Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes. Mol Biol Rep 37, 1125-1135. |
| [9] | Aguilar-Jaramillo AE, Marín-González E, Matías-Hernández L, Osnato M, Pelaz S, Suárez-López P ( 2019). TEMPRANILLO is a direct repressor of the microRNA miR172. Plant J 100, 522-535. |
| [10] | Anwar N, Ohta M, Yazawa T, Sato Y, Li C, Tagiri A, Sakuma M, Nussbaumer T, Bregitzer P, Pourkheirandish M, Wu JZ, Komatsuda T ( 2018). miR172 downregulates the translation of cleistogamy 1 in barley. Ann Bot 122, 251-265. |
| [11] | Aukerman MJ, Sakai H ( 2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730-2741. |
| [12] | Bartel DP ( 2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281-297. |
| [13] | B?umler J, Riber W, Klecker M, Müller L, Dissmeyer N, Weig AR, Mustroph A ( 2019). AtERF#111/ABR1 is a transcriptional activator involved in the wounding response. Plant J 100, 969-990. |
| [14] | Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F ( 2008). MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J 54, 876-887. |
| [15] | Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang LM, Hattori J, Liu CM, van Lammeren AAM, Miki BLA, Custers JBM, van Lookeren Campagne MM ( 2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737-1749. |
| [16] | Bowman JL, Smyth DR, Meyerowitz EM ( 2012). The ABC model of flower development: then and now. Development 139, 4095-4098. |
| [17] | Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O ( 2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185-1190. |
| [18] | Candar-Cakir B, Arican E, Zhang BH ( 2016). Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol J 14, 1727-1746. |
| [19] | Carrington JC, Ambros V ( 2003). Role of microRNAs in plant and animal development. Science 301, 336-338. |
| [20] | Chen XM ( 2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022-2025. |
| [21] | Chuck G, Cigan AM, Saeteurn K, Hake S ( 2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet 39, 544-549. |
| [22] | Chuck G, Meeley R, Hake S ( 2008). Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135, 3013-3019. |
| [23] | Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernié T, Ott T, Gamas P, Crespi M, Niebel A ( 2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20, 3084-3088. |
| [24] | De Luis A, Markmann K, Cognat V, Holt DB, Charpentier M, Parniske M, Stougaard J, Voinnet O ( 2012). Two microRNAs linked to nodule infection and nitrogen-fixing ability in the legume Lotus japonicus. Plant Physiol 160, 2137-2154. |
| [25] | Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K ( 2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33, 751-763. |
| [26] | Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR ( 1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155-168. |
| [27] | Fran?ois L, Verdenaud M, Fu XP, Ruleman D, Dubois A, Vandenbussche M, Bendahmane A, Raymond O, Just J, Bendahmane M ( 2018). A miR172 target-deficient AP2-like gene correlates with the double flower phenotype in roses. Sci Rep 8, 12912. |
| [28] | Frazier TP, Sun GL, Burklew CE, Zhang BH ( 2011). Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol Biotechnol 49, 159-165. |
| [29] | Fu FF, Xue HW ( 2010). Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154, 927-938. |
| [30] | Glazińska P, Zienkiewicz A, Wojciechowski W, Kopcewicz J ( 2009). The putative miR172 target gene InAPETALA2-like is involved in the photoperiodic flower induction of Ipomoea nil. J Plant Physiol 166, 1801-1813. |
| [31] | Han YY, Zhang X, Wang YF, Ming F ( 2013). The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS One 8, e73541. |
| [32] | He YR, Jia RR, Qi JJ, Chen SC, Lei TG, Xu LZ, Peng AH, Yao LX, Long Q, Li ZG, Li Q ( 2019). Functional analysis of citrus AP2 transcription factors identified CsAP2-09 involved in citrus canker disease response and tolerance. Gene 707, 178-188. |
| [33] | Holt DB, Gupta V, Meyer D, Abel NB, Andersen SU, Stougaard J, Markmann K ( 2015). MicroRNA 172 (miR172) signals epidermal infection and is expressed in cells primed for bacterial invasion in Lotus japonicus roots and nodules. New Phytol 208, 241-256. |
| [34] | Houston K, McKim SM, Comadran J, Bonar N, Druka I, Uzrek N, Cirillo E, Guzy-Wrobelska J, Collins NC, Halpin C, Hansson M, Dockter C, Druka A, Waugh R ( 2013). Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc Natl Acad Sci USA 110, 16675-16680. |
| [35] | Hu YX, Wang YX, Liu XF, Li JY ( 2004). Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res 14, 8-15. |
| [36] | Hutvágner G, Zamore PD ( 2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056-2060. |
| [37] | Ji LJ, Liu XG, Yan J, Wang WM, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng BL, Agarwal M, Liu CY, Cao XF, Tang GL, Chen XM ( 2011). ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet 7, e1001358. |
| [38] | Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK ( 1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211-1225. |
| [39] | Jones-Rhoades MW, Bartel DP, Bartel B ( 2006). MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57, 19-53. |
| [40] | José Ripoll J, Bailey LJ, Mai QA, Wu SL, Hon CT, Chapman EJ, Ditta GS, Estelle M, Yanofsky MF ( 2015). MicroRNA regulation of fruit growth. Nat Plants 1, 15036. |
| [41] | Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM ( 2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736-2748. |
| [42] | Khanday I, Skinner D, Yang B, Mercier R, Sundaresan V ( 2019). A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91-95. |
| [43] | Kunst L, Klenz JE, Martinez-Zapater J, Haughn GW ( 1989). AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1, 1195-1208. |
| [44] | Kurihara Y, Watanabe Y ( 2004). Arabidopsis micro-RNA biogenesis through dicer-like 1 protein functions. Proc Natl Acad Sci USA 101, 12753-12758. |
| [45] | Lauter N, Kampani A, Carlson S, Goebel M, Moose SP ( 2005). MicroRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA 102, 9412-9417. |
| [46] | Lee DY, An G ( 2012). Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69, 445-461. |
| [47] | Lee YS, Lee DY, Cho LH, An G ( 2014). Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 7, 31. |
| [48] | Lei M, Li ZY, Wang JB, Fu YL, Xu L ( 2019). Ectopic expression of the Aechmea fasciata APETALA2 gene AfAP2-2 reduces seed size and delays flowering in Arabidopsis. Plant Physiol Biochem 139, 642-650. |
| [49] | Li H, Deng Y, Wu TL, Subramanian S, Yu O ( 2010). Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol 153, 1759-1770. |
| [50] | Li JJ, Yang ZY, Yu B, Liu J, Chen XM ( 2005). Methylation protects miRNAs and siRNAs from a 3°-end uridylation activity in Arabidopsis. Curr Biol 15, 1501-1507. |
| [51] | Li JP, Chen FJ, Li YQ, Li PC, Wang YQ, Mi GH, Yuan LX ( 2019a). ZmRAP2.7, an AP2 transcription factor, is involved in maize brace roots development. Front Plant Sci 10, 820. |
| [52] | Li XY, Guo F, Ma SY, Zhu MY, Pan WH, Bian HW ( 2019b). Regulation of flowering time via miR172-mediated APETALA2-like expression in ornamental gloxinia (Sinningia speciosa). J Zhejiang Univ Sci B 20, 322-331. |
| [53] | Liu P, Liu J, Dong HX, Sun JQ ( 2018). Functional regulation of Q by microRNA172 and transcriptional co-repressor TOPLESS in controlling bread wheat spikelet density. Plant Biotechnol J 16, 495-506. |
| [54] | Llave C, Xie ZX, Kasschau KD, Carrington JC ( 2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053-2056. |
| [55] | Luan YS, Cui J, Li J, Jiang N, Liu P, Meng J ( 2018). Effective enhancement of resistance to Phytophthora infestans by overexpression of miR172a and b in Solanum lycopersicum. Planta 247, 127-138. |
| [56] | Martin A, Adam H, Díaz-Mendoza M, ?urczak M, González-Schain ND, Suárez-López P ( 2009). Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136, 2873-2881. |
| [57] | Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M ( 2009). Repression of flowering by the miR172 target SMZ. PLoS Biol 7, e1000148. |
| [58] | Miao CB, Wang D, He RQ, Liu SK, Zhu JK ( 2020). Mutations in MIR396e and MIR396f increase grain size and modulate shoot architecture in rice. Plant Biotechnol J 18, 491-501. |
| [59] | Miao CB, Wang Z, Zhang L, Yao JJ, Hua K, Liu X, Shi HZ, Zhu JK ( 2019). The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat Commun 10, 3822. |
| [60] | Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K ( 2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819, 86-96. |
| [61] | Mlotshwa S, Yang ZY, Kim Y, Chen XM ( 2006). Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol Biol 61, 781-793. |
| [62] | Moose SP, Sisco PH ( 1994). Glossy15 controls the epidermal juvenile-to-adult phase transition in maize. Plant Cell 6, 1343-1355. |
| [63] | Moose SP, Sisco PH ( 1996). Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev 10, 3018-3027. |
| [64] | Nair SK, Wang N, Turuspekov Y, Pourkheirandish M, Sinsuwongwat S, Chen GX, Sameri M, Tagiri A, Honda I, Watanabe Y, Kanamori H, Wicker T, Stein N, Nagamura Y, Matsumoto T, Komatsuda T ( 2009). Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc Natl Acad Sci USA 107, 490-495. |
| [65] | Nakano T, Suzuki K, Fujimura T, Shinshi H ( 2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140, 411-432. |
| [66] | Nova-Franco B, í?iguez LP, Valdés-López O, Alvarado- Affantranger X, Leija A, Fuentes SI, Ramírez M, Paul S, Reyes JL, Girard L, Hernández G ( 2015). The microRNA172c-APETALA2-1 node as a key regulator of the common bean- Rhizobium etli nitrogen fixation symbiosis. Plant Physiol 168, 273-291. |
| [67] | Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS ( 2005). Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA 102, 3691-3696. |
| [68] | Park W, Li JJ, Song RT, Messing J, Chen XM ( 2002). CARPEL FACTORY, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12, 1484-1495. |
| [69] | Riechmann JL, Meyerowitz EM ( 1998). The AP2/EREBP family of plant transcription factors. Biol Chem 379, 633-646. |
| [70] | Rogers K, Chen XM ( 2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25, 2383-2399. |
| [71] | Sahito ZA, Wang LX, Sun ZX, Yan QQ, Zhang XK, Jiang Q, Ullah I, Tang YP, Li X ( 2017). The miR172c-NNC1 module modulates root plastic development in response to salt in soybean. BMC Plant Biol 17, 229. |
| [72] | Saminathan T, Alvarado A, Lopez C, Shinde S, Gajanayake B, Abburi VL, Vajja VG, Jagadeeswaran G, Raja Reddy K, Nimmakayala P, Reddy UK ( 2019). Elevated carbon dioxide and drought modulate physiology and storage-root development in sweet potato by regulating microRNAs. Funct Integr Genomics 19, 171-190. |
| [73] | Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU ( 2003). Dissection of floral induction pathways using global expression analysis. Development 130, 6001-6012. |
| [74] | Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D ( 2005). Specific effects of microRNAs on the plant transcriptome. Dev Cell 8, 517-527. |
| [75] | Shannon S, Meeks-Wagner DR ( 1993). Genetic interactions that regulate inflorescence development in Arabidopsis. Plant Cell 5, 639-655. |
| [76] | Simons KJ, Fellers JP, Trick HN, Zhang ZC, Tai YS, Gill BS, Faris JD ( 2005). Molecular characterization of the major wheat domestication gene Q. Genetics 172, 547-555. |
| [77] | Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK ( 2006). Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol Biol 61, 897-915. |
| [78] | Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O ( 2008). Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9, 160. |
| [79] | Swaminathan K, Peterson K, Jack T ( 2008). The plant B3 superfamily. Trends Plant Sci 13, 647-655. |
| [80] | Tang MY, Bai X, Niu LJ, Chai X, Chen MS, Xu ZF ( 2018). miR172 regulates both vegetative and reproductive development in the perennial woody plant Jatropha curcas. Plant Cell Physiol 59, 2549-2563. |
| [81] | Wang GD, Xu XP, Wang H, Liu Q, Yang XT, Liao LX, Cai GH ( 2019a). A tomato transcription factor, SlDREB3 enhances the tolerance to chilling in transgenic tomato. Plant Physiol Biochem 142, 254-262. |
| [82] | Wang L, Sun SY, Jin JY, Fu DB, Yang XF, Weng XY, Xu CG, Li XH, Xiao JH, Zhang QF ( 2015a). Coordinated regulation of vegetative and reproductive branching in rice. Proc Natl Acad Sci USA 112, 15504-11509. |
| [83] | Wang TY, Ping XK, Cao YR, Jian HJ, Gao YM, Wang J, Tan YC, Xu XF, Lu K, Li JN, Liu LZ ( 2019b). Genome-wide exploration and characterization of miR172/ euAP2 genes in Brassica napus L. for likely role in flower organ development. BMC Plant Biol 19, 336. |
| [84] | Wang Y, Wang ZS, Amyot L, Tian LN, Xu ZQ, Gruber MY, Hannoufa A ( 2015b). Ectopic expression of miR156 represses nodulation and causes morphological and developmental changes in Lotus japonicus. Mol Genet Genomics 290, 471-484. |
| [85] | Wang YN, Li KX, Chen L, Zou YM, Liu HP, Tian YP, Li DX, Wang R, Zhao F, Ferguson BJ, Gresshoff PM, Li X (c). 2015 MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol 168, 984-999. |
| [86] | Wang YN, Wang LX, Zou YM, Chen L, Cai ZM, Zhang SL, Zhao F, Tian YP, Jiang Q, Ferguson BJ, Gresshoff PM, Li X ( 2014). Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26, 4782-4801. |
| [87] | Wang YW, Li PC, Cao XF, Wang XJ, Zhang AM, Li X ( 2009). Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem Biophys Res Commun 378, 799-803. |
| [88] | Wollmann H, Mica E, Todesco M, Long JA, Weigel D ( 2010). On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633-3642. |
| [89] | Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS ( 2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750-759. |
| [90] | Xie ZL, Nolan TM, Jiang H, Yin YH ( 2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front Plant Sci 10, 228. |
| [91] | Yan Z, Hossain MS, Wang J, Valdés-López O, Liang Y, Libault M, Qiu LJ, Stacey G ( 2013). miR172 regulates soybean nodulation. Mol Plant Microbe Interact 26, 1371-1377. |
| [92] | Yang JW, Zhang N, Ma CY, Qu Y, Si HJ, Wang D ( 2013). Prediction and verification of microRNAs related to proline accumulation under drought stress in potato. Comput Biol Chem 46, 48-54. |
| [93] | Zhang KX, Zhao L, Yang X, Li MM, Sun JZ, Wang K, Li YH, Zheng YH, Yao YH, Li WB ( 2019). GmRAV1 regulates regeneration of roots and adventitious buds by the cytokinin signaling pathway in Arabidopsis and soybean. Physiol Plant 165, 814-829. |
| [94] | Zhao XH, Cao D, Huang ZJ, Wang JL, Lu SJ, Xu Y, Liu BH, Kong FJ, Yuan XH ( 2015). Dual functions of GmTOE4a in the regulation of photoperiod-mediated flowering and plant morphology in soybean. Plant Mol Biol 88, 343-355. |
| [95] | Zhao XY, Qi CH, Jiang H, You CX, Guan QM, Ma FW, Li YY, Hao YJ ( 2019). The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Hortic Res 6, 66. |
| [96] | Zhou XF, Wang GD, Sutoh K, Zhu JK, Zhang WX ( 2008). Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim Biophys Acta 1779, 780-788. |
| [97] | Zhu QH, Helliwell CA ( 2011). Regulation of flowering time and floral patterning by miR172. J Exp Bot 62, 487-495. |
| [98] | Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA ( 2009). Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice ( Oryza sativa). BMC Plant Biol 9, 149. |
| [99] | Zou YM, Wang YN, Wang LX, Yang L, Wang R, Li X ( 2013). miR172b controls the transition to autotrophic development inhibited by ABA in Arabidopsis. PLoS One 8, e64770. |