

植物DREPP基因家族研究进展
收稿日期: 2019-01-21
录用日期: 2019-04-23
网络出版日期: 2019-04-29
基金资助
黑龙江省自然科学基金(2019001001);中央高校基本科研业务费专项(No.2572017ET01);中央高校基本科研业务费专项(No.2572018BS03)
Research Advances in DREPP Gene Family in Plants
Received date: 2019-01-21
Accepted date: 2019-04-23
Online published: 2019-04-29
张洵 , 喻娟娟 , 王思竹 , 李莹 , 戴绍军 . 植物DREPP基因家族研究进展[J]. 植物学报, 2019 , 54(5) : 582 -595 . DOI: 10.11983/CBB19014
Developmentally regulated plasma membrane polypeptide (DREPP) proteins, a family of plant-specific proteins associated with the plasma membrane, have multiple functions such as combining PtdInsPs, the Ca 2+/CaM complex, microtubules and microfilaments. DREPPs play an important role in plant growth and development and response to stress (e.g., low temperature and drought). This paper reviews the composition of the DREPP family as well as their protein sequence characteristics and biological functions during development and stress response and provides information for understanding how DREPPs mediate signaling networks.
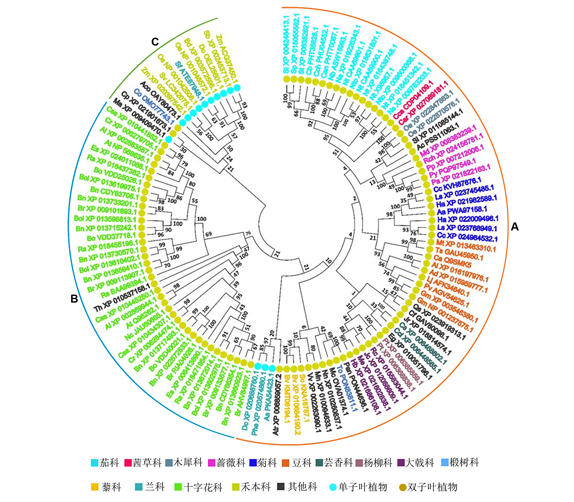
Key words: DREPP; plant development; abiotic stress
| 1 | 刘焱, 邢立静, 李俊华, 戴绍军 (2012). 水稻含有B-box锌指结构域的OsBBX25蛋白参与植物对非生物胁迫的响应. 植物学报 47, 366-378. |
| 2 | 邱丽丽, 赵琪, 张玉红, 戴绍军 (2017). 植物质膜蛋白质组的逆境应答研究进展. 植物学报 52, 128-147. |
| 3 | 喻娟娟, 戴绍军 (2009). 植物蛋白质组学研究若干重要进展. 植物学报 44, 410-425. |
| 4 | Alexandrov NN, Brover VV, Freidin S, Troukhan ME, Tatarinova TV, Zhang HY, Swaller TJ, Lu YP, Bouck J, Flavell RB, Feldmann KA (2009). Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol Biol 69, 179-194. |
| 5 | Buschmann H, Lloyd CW (2008). Arabidopsis mutants and the network of microtubule-associated functions. Mol Plant 1, 888-898. |
| 6 | Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438, 1013-1016. |
| 7 | Dohm JC, Minoche AE, Holtgr?we D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, S?rensen TR, Stracke R, Reinhardt R, Goesmann A, Kraft T, Schulz B, Stadler PF, Schmidt T, Gabaldón T, Lehrach H, Weisshaar B, Himmelbauer H (2014). The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 23, 546-549. |
| 8 | Fleta-Soriano E, Munné-Bosch S (2016). Stress memory and the inevitable effects of drought: a physiological perspective. Front Plant Sci 7, 143. |
| 9 | Fukushima K, Fang XD, Alvarez-Ponce D, Cai HM, Carretero-Paulet L, Chen C, Chang TH, Farr KM, Fujita T, Hiwatashi Y, Hoshi Y, Imai T, Kasahara M, Librado P, Mao LK, Mori H, Nishiyama T, Nozawa M, Pálfalvi G, Pollard ST, Rozas J, Sánchez-Gracia A, Sankoff D, Shibata TF, Shigenobu S, Sumikawa N, Uzawa T, Xie MY, Zheng CF, Pollock DD, Albert VA, Li SC, Hasebe M (2017). Genome of the pitcher plant Cephalotus reveals genetic changes associated with carnivory. Nat Ecol Evol 1, 59. |
| 10 | Gantet P, Masson F, Domergue O, Marquis-Mention M, Bauw G, Inze D, Rossignol M, de la Serve BT (1996). Cloning of a cDNA encoding a developmentally regulated 22 kDa polypeptide from tobacco leaf plasma membrane. Biochem Mol Biol Int 40, 469-477. |
| 11 | Huang YP, Huang YW, Chen IH, Shenkwen LL, Hsu YH, Tsai CH (2017). Plasma membrane-associated cation- binding protein 1-like protein negatively regulates inter-cellular movement of BaMV. J Exp Bot 68, 4765-4774. |
| 12 | Hunt L, Otterhag L, Lee JC, Lasheen T, Hunt J, Seki M, Shinozaki K, Sommarin M, Gilmour DJ, Pical C, Gray JE (2004). Gene-specific expression and calcium activation of Arabidopsis thaliana phospholipase C isoforms. New Phytol 162, 643-654. |
| 13 | Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000). Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24, 655-665. |
| 14 | Ide Y, Nagasaki N, Tomioka R, Suito M, Kamiya T, Maeshima M (2007). Molecular properties of a novel, hydrophilic cation-binding protein associated with the plasma membrane. J Exp Bot 58, 1173-1183. |
| 15 | Jones MA, Shen JJ, Fu Y, Li H, Yang ZB, Grierson CS (2002). The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 14, 763-776. |
| 16 | Kaadige MR, Ayer DE (2006). The polybasic region that follows the plant homeodomain zinc finger 1 of Pf1 is necessary and sufficient for specific phosphoinositide binding. J Biol Chem 281, 28831-28836. |
| 17 | Kang EF, Zheng MZ, Zhang Y, Yuan M, Yalovsky S, Zhu L, Fu Y (2017). The microtubule-associated protein MAP18 affects ROP2 GTPase activity during root hair growth. Plant Physiol 174, 202-222. |
| 18 | Kato M, Aoyama T, Maeshima M (2013). The Ca 2+-binding protein PCaP2 located on the plasma membrane is involved in root hair development as a possible signal transducer . Plant J 74, 690-700. |
| 19 | Kato M, Nagasaki-Takeuchi N, Ide Y, Maeshima M (2010a). An Arabidopsis hydrophilic Ca 2+-binding protein with a PEVK-rich domain, PCaP2, is associated with the plasma membrane and interacts with calmodulin and phosphatidylinositol phosphates. Plant Cell Physiol 51, 366-379. |
| 20 | Kato M, Nagasaki-Takeuchi N, Ide Y, Tomioka R, Mae- shima M (2010b). PCaPs, possible regulators of PtdInsP signals on plasma membrane. Plant Signal Behav 5, 848-850. |
| 21 | Kaur P, Appels R, Bayer PE, Keeble-Gagnere G, Wang JK, Hirakawa H, Shirasawa K, Vercoe P, Stefanova K, Durmic Z, Nichols P, Revell C, Isobe SN, Edwards D, Erskine W (2017). Climate clever clovers: new paradigm to reduce the environmental footprint of ruminants by breeding low methanogenic forages utilizing haplotype variation. Front Plant Sci 8, 1463. |
| 22 | Kim S, Park J, Yeom SI, Kim YM, Seo E, Kim KT, Kim MS, Lee JM, Cheong K, Shin HS, Kim SB, Han K, Lee J, Park M, Lee HA, Lee HY, Lee Y, Oh S, Lee JH, Choi E, Choi E, Lee SE, Jeon J, Kim H, Choi G, Song H, Lee J, Lee SC, Kwon JK, Lee HY, Koo N, Hong YJ, Kim RW, Kang WH, Huh JH, Kang BC, Yang TJ, Lee YH, Bennetzen JL, Choi D (2017). New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol 18, 210. |
| 23 | Kost B (2008). Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol 18, 119-127. |
| 24 | Lee YJ, Yang ZB (2008). Tip growth: signaling in the apical dome. Curr Opin Plant Biol 11, 662-671. |
| 25 | Li JJ, Wang XL, Qin T, Zhang Y, Liu XM, Sun JB, Zhou Y, Zhu L, Zhang ZD, Yuan M, Mao TL (2011). MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. Plant Cell 23, 4411-4427. |
| 26 | Liu WX, Zhang FC, Zhang WZ, Song LF, Wu WH, Chen YF (2013). Arabidopsis Di19 functions as a transcription factor and modulates PR1, PR2, and PR5 expression in response to drought stress. Mol Plant 6, 1487-1502. |
| 27 | Liu XB, Liu YS, Huang P, Ma YS, Qing ZX, Tang Q, Cao HF, Cheng P, Zheng YJ, Yuan ZJ, Zhou Y, Liu JF, Tang ZS, Zhuo YX, Zhang YC, Yu LL, Huang JL, Yang P, Peng Q, Zhang JB, Jiang WK, Zhang ZH, Lin K, Ro DK, Chen XY, Xiong XY, Shang Y, Huang SW, Zeng JG (2017). The genome of medicinal plant Macleaya cordata provides new insights into benzylisoquinoline alkaloids metabolism. Mol Plant 10, 975-989. |
| 28 | Logan DC, Domergue O, de la Serve B T, Rossignol M (1998). A new family of plasma membrane polypeptides differentially regulated during plant development. Biochem Mol Biol Int 43, 1051-1062. |
| 29 | Mao J, Liu Q, Yang X, Long C, Zhao M, Zeng H, Liu H, Yuan J, Qiu D (2010). Purification and expression of a protein elicitor from Alternaria tenuissima and elicitor- mediated defence responses in tobacco. Ann App Biol 156, 411-420. |
| 30 | Matthews BF, Beard H, MacDonald MH, Kabir S, Youssef RM, Hosseini P, Brewer E (2013). Engineered resistance and hypersusceptibility through functional metabolic studies of 100 genes in soybean to its major pathogen, the soybean cyst nematode. Planta 5, 1337-1357. |
| 31 | Meng FL, Xiao Y, Guo LH, Zeng HM, Yang XF, Qiu DW (2018). A DREPP protein interacted with PeaT1 from Alternaria tenuissima and is involved in elicitor-induced disease resistance in Nicotiana plants. J Plant Res 131, 827-837. |
| 32 | Nagasaki-Takeuchi N, Miyano M, Maeshima M (2008). A plasma membrane-associated protein of Arabidopsis thaliana AtPCaP1 binds copper ions and changes its higher order structure. J Biochem 144, 487-497. |
| 33 | Novillo F, Alonso JM, Ecker JR, Salinas J (2004). CBF2/ DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 101, 3985-3990. |
| 34 | Novillo F, Medina J, Salinas J (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc Natl Acad Sci USA 104, 21002-21007. |
| 35 | Qin T, Liu XM, Li JJ, Sun JB, Song LN, Mao TL (2014). Arabidopsis microtubule-destabilizing protein 25 functions in pollen tube growth by severing actin filaments. Plant Cell 26, 325-339. |
| 36 | Redwan RM, Saidin A, Kumar SV (2016). The draft genome of MD-2 pineapple using hybrid error correction of long reads. DNA Res 23, 427-439. |
| 37 | Scaglione D, Reyes-Chin-Wo S, Acquadro A, Froenicke L, Portis E, Beitel C, Tirone M, Mauro R, Lo Monaco A, Mauromicale G, Faccioli P, Cattivelli L, Rieseberg L, Michelmore R, Lanteri S (2016). The genome sequence of the outbreeding globe artichoke constructed de novo incorporating a phase-aware low-pass sequencing stra-tegy of F1 progeny. Sci Rep 6, 19427. |
| 38 | Shen Q, Zhang LD, Liao ZH, Wang SY, Yan TX, Shi P, Liu M, Fu XQ, Pan QF, Wang YL, Lv ZY, Lu X, Zhang FY, Jiang WM, Ma YA, Chen MH, Hao XL, Li L, Tang YL, Lv G, Zhou Y, Sun XF, Brodelius PE, Rose JKC, Tang KX (2018). The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol Plant 11, 776-788. |
| 39 | Shi YT, Ding Yl, Yang SH (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci 23, 623-637. |
| 40 | Studer AJ, Schnable JC, Weissmann S, Kolbe AR, McKain MR, Shao Y, Cousins AB, Kellogg EA, Brutnell TP (2016). The draft genome of the C3 panicoid grass species Dichanthelium oligosanthes. Genome Biol 17, 223. |
| 41 | Subba P, Barua P, Kumar R, Datta A, Soni KK, Chakraborty S, Chakraborty N (2013). Phosphoproteomic dynamics of chickpea ( Cicer arietinum L.) reveals shared and distinct components of dehydration response. J Proteome Res 12, 5025-5047. |
| 42 | Testerink C, Munnik T (2005). Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci 10, 368-375. |
| 43 | Theerawitaya C, Yamada-Kato N, Singh HP, Cha-Um S, Takabe T (2018). Isolation, expression, and functional analysis of developmentally regulated plasma membrane polypeptide 1 (DREPP1) in Sporobolus virginicus grown under alkali salt stress. Protoplasma 255, 1423-1432. |
| 44 | Trivedi DK, Gill SS, Tuteja N (2016). Abscisic acid (ABA): biosynthesis, regulation, and role in abiotic stress tole-rance. In: Tuteja N, Gill SS, eds. Abiotic Stress Response in Plants. Weinheim: Wiley Wiley-VCH Verlag GmbH & Co. pp. 311-322. |
| 45 | Vosolsobě S, Petrá?ek J, Schwarzerová K (2017). Evolutionary plasticity of plasma membrane interaction in DREPP family proteins. Biochim Biophys Acta Biomembr 1859, 686-697. |
| 46 | Wang X, Zhu L, Liu BQ, Wang C, Jin LF, Zhao Q, Yuan M (2007). Arabidopsis MICROTUBULE-ASSOCIATED PRO- TEIN 18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19, 877-889. |
| 47 | Wang XL, Wang L, Wang Y, Liu H, Hu D, Zhang N, Zhang SB, Cao HY, Cao QY, Zhang ZH, Tang S, Song DD, Wang C (2018a) . Arabidopsis PCaP2 plays an important role in chilling tolerance and ABA response by activating CBF- and SnRK2-mediated transcriptional regulatory network. Front Plant Sci 9, 215. |
| 48 | Wang XL, Wang Y, Wang L, Liu H, Zhang B, Cao QJ, Liu XY, Bi ST, Lv YL, Wang QY, Zhang SB, He M, Tang S, Yao S, Wang C (2018b). Arabidopsis PCaP2 functions as a linker between ABA and SA signals in plant water deficit tolerance. Front Plant Sci 9, 578. |
| 49 | Yamada N, Theerawitaya C, Kageyama H, Cha-Um S, Takabe T (2015). Expression of developmentally regulated plasma membrane polypeptide (DREPP2) in rice root tip and interaction with Ca 2+/CaM complex and microtubule . Protoplasma 252, 1519-1527. |
| 50 | Yu JJ, Zhang YX, Liu JM, Wang L, Liu PP, Yin ZP, Guo SY, Ma J, Lu Z, Wang T, She YM, Miao YC, Ma L, Chen SX, Li Y, Dai SJ (2018). Proteomic discovery of H2O2 response in roots and functional characterization of PutGLP gene from alkaligrass. Planta 248, 1079-1099. |
| 51 | Yuasa K, Maeshima M (2000). Purification, properties, and molecular cloning of a novel Ca 2+-binding protein in radish vacuoles . Plant Physiol 124, 1069-1078. |
| 52 | Zhang Q, Qu YN, Jing W, Li L, Zhang WH (2014). Phospholipase Ds in plant response to hyperosmotic stresses. In: Wang XM, ed. Phospholipases in Plant Signaling. Berlin, Heidelberg: Springer. pp. 121-134. |
| 53 | Zhang X, Wei LQ, Wang ZZ, Wang T (2013). Physiological and molecular features of Puccinellia tenuiflora tolerating salt and alkaline-salt stress. J Integr Plant Biol 55, 262-276. |
| 54 | Zhao Q, Suo JW, Chen SX, Jin YD, Ma XL, Yin ZP, Zhang YH, Wang T, Luo J, Jin WH, Zhang X, Zhou ZQ, Dai SJ (2016). Na2CO3-responsive mechanisms in halophyte Puccinellia tenuiflora roots revealed by physiological and proteomic analyses. Sci Rep 6, 32717. |
| 55 | Zhu JK (2016). Abiotic stress signaling and responses in plants. Cell 167, 313-324. |
| 56 | Zhu L, Zhang Y, Kang EF, Xu QY, Wang MY, Rui Y, Liu BQ, Yuan M, Fu Y (2013). MAP18 regulates the direction of pollen tube growth in Arabidopsis by modulating F-actin organization. Plant Cell 25, 851-867. |
/
| 〈 |
|
〉 |