

单细胞组学技术及其在植物保卫细胞研究中的应用
收稿日期: 2016-08-08
录用日期: 2017-03-06
网络出版日期: 2017-03-06
基金资助
河南省自然科学基金(No.162300410008)和河南省高等学校重点科研项目(No.15A180012)
Applications of Single-cell Technologies in Guard Cells
Received date: 2016-08-08
Accepted date: 2017-03-06
Online published: 2017-03-06
牛艳丽 , 柏胜龙 , 王麒云 , 刘凌云 . 单细胞组学技术及其在植物保卫细胞研究中的应用[J]. 植物学报, 2017 , 52(6) : 788 -796 . DOI: 10.11983/CBB16164
Single-cell technologies have been used in an increasing number of animal studies, but the techniques have yet to be widely used in plants, especially in guard cells. Stomatal pores are formed by pairs of guard cells and serve as major gateways for both CO2 influx into plants from the atmosphere and transpirational water loss. The application of single-cell technologies will be valuable for better understanding the underlying mechanisms of stomatal pores. In this review, we discuss single-cell technologies, current research and problems with guard cells and focus on the application of single-cell technologies to guard cells. Single-cell technologies may have potential to provide new perspectives for plant problems, such as development, metabolism and response to environmental stresses of guard cells.
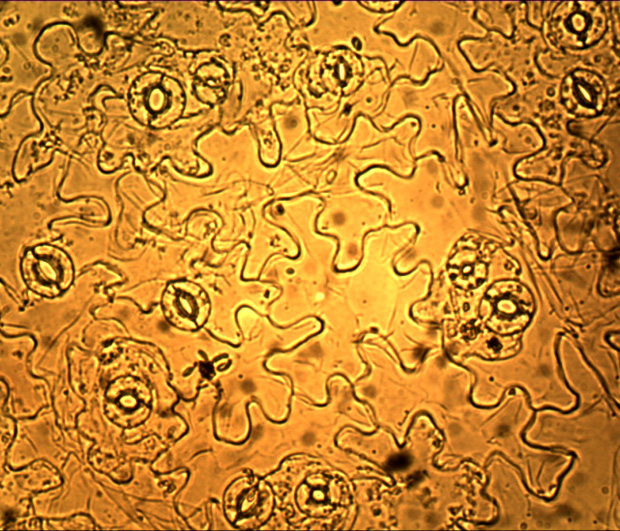
Key words: single cell; guard cells; functional analysis; signaling
| [1] | 孟繁霞, 张蜀秋, 娄成后 (2000). 气孔功能的结构基础. 植物学通报 17, 27-33. |
| [2] | Adrian J, Chang J, Ballenger CE, Bargmann BOR, Alassimone J, Davies KA, Lau OS, Matos JL, Hachez C, Lanctot A, Vatén A, Birnbaum KD, Bergmann DC (2015). Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing pop- ulation.Dev Cell 33, 107-118. |
| [3] | Anjam MS, Ludwig Y, Hochholdinger F, Miyaura C, Inada M, Siddique S, Grundler FMW (2016). An improved procedure for isolation of high-quality RNA from nematode- infected Arabidopsis roots through laser capture microdissection.Plant Methods 12, 25. |
| [4] | Aubry S, Aresheva O, Reyna-Llorens I, Smith-Unna RD, Hibberd JM, Genty B (2016). A specific transcriptome signature for guard cells from the C4 plant Gynandropsis gynandra. Plant Physiol 170, 1345-1357. |
| [5] | Barkla BJ, Vera-Estrella R (2015). Single cell-type compara- tive metabolomics of epidermal bladder cells from the hal- ophyte Mesembryanthemum crystallinum. Front Plant Sci 6, 435. |
| [6] | Barkla BJ, Vera-Estrella R, Raymond C (2016). Single-cell- type quantitative proteomic and ionomic analysis of epi- dermal bladder cells from the halophyte model plant Mes- embryanthemum crystallinum to identify salt-responsive proteins. BMC Plant Biol 16, 110. |
| [7] | Cápal P, Blavet N, Vrána J, Kubaláková M, Dole?el J (2015). Multiple displacement amplification of the DNA from single flow-sorted plant chromosome.Plant J 84, 838-844. |
| [8] | Carter AD, Bonyadi R, Gifford ML (2013). The use of fluorescence-activated cell sorting in studying plant development and environmental responses.Int J Dev Biol 57, 545-552. |
| [9] | Dean FB, Hosono S, Fang LH, Wu XH, Faruqi AF, Bray- Ward P, Sun ZY, Zong QL, Du FY, Du J, Driscoll M, Song WM, Kingsmore SF, Egholm M, Lasken RS (2002). Comprehensive human genome amplification using multiple displacement amplification.Proc Natl Acad Sci USA 99, 5261-5266. |
| [10] | Deng CL, Bai LL, Li SF, Zhang YX, Li X, Chen YH, Wang RRC, Han FP, Hu ZM (2014). DOP-PCR based painting of rye chromosomes in a wheat background.Genome 57, 473-479. |
| [11] | Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ (2004). ABA, hydrogen peroxide and nitric oxide signaling in stomatal guard cells.J Exp Bot 55, 205-212. |
| [12] | Dow GJ, Bergmann DC, Berry JA (2014). An integrated model of stomatal development and leaf physiology.New Phytol 201, 1218-1226. |
| [13] | Efroni I, Birnbaum KD (2016). The potential of single-cell profiling in plants.Genome Biol 17, 65. |
| [14] | Efroni I, Mello A, Nawy T, Ip PL, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD (2016). Root regeneration triggers an embryo-like sequence guided by hormonal interactions.Cell 165, 1721-1733. |
| [15] | Fritzsch FS, Dusny C, Frick O, Schmid A (2012). Single-cell analysis in biotechnology, systems biology, and biocatalysis.Annu Rev Chem Biomol Eng 3, 129-155. |
| [16] | Fu YS, Li CM, Lu SJ, Zhou WX, Tang FC, Xie XS, Huang YY (2015). Uniform and accurate single-cell sequencing based on emulsion whole-genome amplification.Proc Natl Acad Sci USA 112, 11923-11928. |
| [17] | Fujii T, Matsuda S, Tejedor ML, Esaki T, Sakane I, Mizuno H, Tsuyama N, Masujima T (2015). Direct metabolomics for plant cells by live single-cell mass spectrometry.Nat Protoc 10, 1445-1456. |
| [18] | Grasso MS, Lintilhac PM (2016). Microbead encapsulation of living plant protoplasts: a new tool for the handling of single plant cells.Appl Plant Sci 4, 1500140. |
| [19] | Grondin A, Rodrigues O, Verdoucq L, Merlot S, Leonhardt N, Maurel C (2015). Aquaporins contribute to ABA- triggered stomatal closure through OST1-mediated phosphorylation.Plant Cell 27, 1945-1954. |
| [20] | Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R, Koltay P (2015). Technologies for single-cell isolation.Int J Mol Sci 16, 16897-16919. |
| [21] | Guillaume-Gentil O, Grindberg RV, Kooger R, Dorwling- Carter L, Martinez V, Ossola D, Pilhofer M, Zambelli T, Vorholt JA (2016). Tunable single-cell extraction for mole- cular analyses.Cell 166, 506-516. |
| [22] | Harada E, Kim JA, Meyer AJ, Hell R, Clemens S, Choi YE (2010). Expression profiling of tobacco leaf trichomes iden- tifies genes for biotic and abiotic stresses.Plant Cell Phy- siol 51, 1627-1637. |
| [23] | He JM, Ma XG, Zhang Y, Sun TF, Xu FF, Chen YP, Liu X, Yue M (2013). Role and interrelationship of Gα protein, hydrogen peroxide, and nitric oxide in ultraviolet B-induced stomatal closure in Arabidopsis leaves.Plant Physiol 161, 1570-1583. |
| [24] | Hossain MS, Joshi T, Stacey G (2015). System approaches to study root hairs as a single cell plant model: current status and future perspectives.Front Plant Sci 6, 363. |
| [25] | Hou Y, Guo HH, Cao C, Li XL, Hu BQ, Zhu P, Wu XL, Wen L, Tang FC, Huang YY, Peng JR (2016). Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas.Cell Res 26, 304-319. |
| [26] | Huang L, Ma F, Chapman A, Lu SJ, Xie XS (2015). Single- cell whole-genome amplification and sequencing: methodo- logy and applications.Annu Rev Genomics Hum Genet 16, 79-102. |
| [27] | Huang XH, Wei XH, Sang T, Zhao Q, Feng Q, Zhao Y, Li CY, Zhu CR, Lu TT, Zhang ZW, Li M, Fan DL, Guo YL, Wang AH, Wang L, Deng LW, Li WJ, Lu YQ, Weng QJ, Liu KY, Huang T, Zhou TY, Jing YF, Li W, Lin Z, Buckler ES, Qian Q, Zhang QF, Li JY, Han B (2010). Genome-wide association studies of 14 agronomic traits in rice landraces.Nat Genet 42, 961-967. |
| [28] | Huang XH, Yang SH, Gong JY, Zhao Y, Feng Q, Gong H, Li WJ, Zhan QL, Cheng BY, Xia JH, Chen N, Hao ZN, Liu KY, Zhu CR, Huang T, Zhao Q, Zhang L, Fan DL, Zhou CC, Lu YQ, Weng QJ, Wang ZX, Li JJ, Han B (2015). Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis.Nat Commun 6, 6258. |
| [29] | Iyer-Pascuzzi AS, Benfey PN (2010). Fluorescence activa- ted cell sorting in plant developmental biology.Methods Mol Biol 655, 313-319. |
| [30] | Jin XF, Wang RS, Zhu MM, Jeon BW, Albert R, Chen SX, Assmann SM (2013). Abscisic acid-responsive guard cell metabolomes of Arabidopsis wild-type and gpa1 G-protein mutants. Plant Cell 25, 4789-4811. |
| [31] | Joudoi T, Shichiri Y, Kamizono N, Akaike T, Sawa T, Yoshitake J, Yamada N, Iwai S (2013). Nitrated cyclic GMP modulates guard cell signaling in Arabidopsis.Plant Cell 25, 558-571. |
| [32] | Kehr J (2003). Single cell technology.Curr Opin Plant Biol 6, 617-621. |
| [33] | Lau OS, Davies KA, Chang J, Adrian J, Rowe MH, Ballenger CE, Bergmann DC (2014). Direct roles of SPEECHLESS in the specification of stomatal self-rene- wing cells.Science 345, 1605-1609. |
| [34] | Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abs- cisic acid hypersensitive protein phosphatase 2C mutant.Plant Cell 16, 596-615. |
| [35] | Li X, Li L, Yan JB (2015). Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize.Nat Commun 6, 6648. |
| [36] | Ludwig Y, Hochholdinger F (2014). Laser microdissection of plant cells.Methods Mol Biol 1080, 249-258. |
| [37] | Maisch J, Kreppenhofer K, Büchler S, Merle C, Sobich S, G?rling B, Luy B, Ahrens R, Guber AE, Nick P (2016). Time-resolved NMR metabolomics of plant cells based on a microfluidic chip.J Plant Physiol 200, 28-34. |
| [38] | Matrosova A, Bogireddi H, Mateo-Pe?as A, Hashimoto- Sugimoto M, Iba K, Schroeder JI, Israelsson-Nord- str?m M (2015). The HT1 protein kinase is essential for red light-induced stomatal opening and genetically inter- acts with OST1 in red light and CO2 -induced stomatal movement responses.New Phytol 208, 1126-1137. |
| [39] | Misra BB, Acharya BR, Granot D, Assmann SM, Chen SX (2015a). The guard cell metabolome: functions in stomatal movement and global food security.Front Plant Sci 6, 334. |
| [40] | Misra BB, Assmann SM, Chen SX (2014). Plant single-cell and single-cell-type metabolomics.Trends Plant Sci 19, 637-646. |
| [41] | Misra BB, de Armas E, Tong ZH, Chen SX (2015b). Metabolomic responses of guard cells and mesophyll cells to Bicarbonate.PLoS One 10, e0144206. |
| [42] | Mustroph A, Zanetti ME, Girke T, Bailey-Serres J (2013). Isolation and analysis of mRNAs from specific cell types of plants by ribosome immunopurification.Methods Mol Biol 959, 277-302. |
| [43] | Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis.Proc Natl Acad Sci USA 106, 18843-18848. |
| [44] | Ou XB, Gan Y, Chen PL, Qiu MQ, Jiang K, Wang GX (2014). Stomata prioritize their responses to multiple biotic and abiotic signal inputs.PLoS One 9, e101587. |
| [45] | Pillitteri LJ, Torii KU (2012). Mechanisms of stomatal development.Annu Rev Plant Biol 63, 591-614. |
| [46] | Schad M, Mungur R, Fiehn O, Kehr J (2005). Metabolic profiling of laser microdissected vascular bundles ofArabi- dopsis thaliana. Plant Methods 1, 2. |
| [47] | Sturtevant D, Lee YJ, Chapman KD (2016). Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ. Curr Opin Biotechnol 37, 53-60. |
| [48] | Sun QX, Liu J, Zhang Q, Qing XH, Dobson G, Li XZ, Qi BX (2013). Characterization of three novel desaturases involved in the delta-6 desaturation pathways for polyunsaturated fatty acid biosynthesis from Phytophthora infestans. Appl Microbiol Biotechnol 97, 7689-7697. |
| [49] | Telenius H, Carter NP, Bebb CE, Nordenskj?ld M, Ponder BAJ, Tunnacliffe A (1992). Degenerate oligonucleotide- primed PCR: general amplification of target DNA by a single degenerate primer.Genomics 13, 718-725. |
| [50] | Thakare D, Yang RL, Steffen JG, Zhan JP, Wang DF, Clark RM, Wang XF, Yadegari R (2014). RNA-seq analysis of laser-capture microdissected cells of the developing central starchy endosperm of maize.Genom Data 2, 242-245. |
| [51] | Wang RS, Pandey S, Li S, Gookin TE, Zhao ZX, Albert R, Assmann SM (2011). Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells.BMC Genomics 12, 216. |
| [52] | Yang YZ, Costa A, Leonhardt N, Siegel RS, Schroeder JI (2008). Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool.Plant Methods 4, 6. |
| [53] | Zhao ZX, Stanley BA, Zhang W, Assmann SM (2010). ABA-regulated G protein signaling in Arabidopsis guard cells: a proteomic perspective.J Proteome Res 9, 1637-1647. |
| [54] | Zhao ZX, Zhang W, Stanley BA, Assmann SM (2008). Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20, 3210-3226. |
| [55] | Zhu MM, Dai SJ, McClung S, Yan XF, Chen SX (2009). Functional differentiation of Brassica napus guard cells and mesophyll cells revealed by comparative proteomics. Mol Cell Proteomics 8, 752-766. |
| [56] | Zhu YD, Li H, Bhatti S, Zhou SP, Yang Y, Fish T, Thannhauser TW (2016). Development of a laser capture microscope-based single-cell-type proteomics tool for study- ing proteomes of individual cell layers of plant roots.Hortic Res 3, 16026. |
| [57] | Zong CH, Lu SJ, Chapman AR, Xie XS (2012). Genome- wide detection of single-nucleotide and copy-number variations of a single human cell.Science 338, 1622-1626. |
/
| 〈 |
|
〉 |