

植物颗粒结合淀粉合酶GBSS基因家族的进化
收稿日期: 2016-03-08
录用日期: 2016-08-08
网络出版日期: 2017-04-05
基金资助
国家自然科学基金(No.31570218)
Evolutionary Pattern of the GBSS Gene Family in Plants
Received date: 2016-03-08
Accepted date: 2016-08-08
Online published: 2017-04-05
孙文静 , 包颖 , 王倩 . 植物颗粒结合淀粉合酶GBSS基因家族的进化[J]. 植物学报, 2017 , 52(2) : 179 -187 . DOI: 10.11983/CBB16041
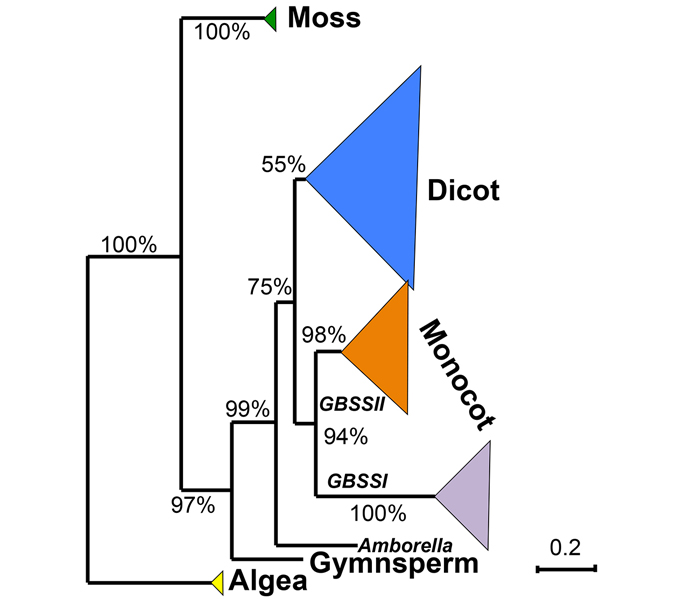
We analyzed the evolutionary pattern of the granule-bound starch-synthase genes (GBSS) in 20 land plants and 2 algae from whole-genome data with bioinformatics methods. A total of 42 genes were detected and their sequence structures were characterized. Phylogenetic analysis revealed that the GBSS gene family had an ancient origin and complex evolutionary history. It probably appeared in the early evolutionary stage of green plants, then mostly experienced lineage-specific expansion and copy loss during evolution, and finally was fixed in different plant taxa via functional divergence.
Key words: genome; phylogeny; gene duplication,; copy loss; functional; divergence
| [1] | 包颖, 杜家潇, 景翔, 徐思 (2015). 药用野生稻叶中淀粉合成酶基因家族的序列分化和特异表达. 植物学报50, 683-690. |
| [2] | Ahuja G, Jaiswal S, Hucl P, Chibbar RN (2014). Wheat genome specific granule-bound starch synthase I differentially influence grain starch synthesis.Carbohydr Polym 114, 87-94. |
| [3] | Alison MS (2012). Starch in the Arabidopsis plant.Starch 64, 421-434. |
| [4] | Ball S, Colleoni C, Cenci U, Raj JN, Tirtiaux C (2011). The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis.J Exp Bot 62, 1775-1801. |
| [5] | Baranov Iu O, Slishchuk HI, Volkova NE, SyvolapIu M (2014). Bioinformatic analysis of maize granule-bound starch synthase gene.Tsitol Genet 48, 18-23. |
| [6] | Criscuolo A (2011). morePhyML: improving the phylogenetic tree space exploration with PhyML 3.Mol Phylogenet Evol 61, 944-948. |
| [7] | Deschamps P, Moreau H, Worden AZ, Dauvillee D, Ball SG (2008). Early gene duplication within chloroplastida and its correspondence with relocation of starch metabolism to chloroplasts.Genetics 178, 2373-2387. |
| [8] | Dian W, Jiang H, Chen Q, Liu F, Wu P (2003). Cloning and characterization of the granule-bound starch synthase II gene in rice: gene expression is regulated by the nitrogen level, sugar and circadian rhythm.Planta 218, 261-268. |
| [9] | Fasahat P, Rahman S, Ratnam W (2014). Genetic controls on starch amylose content in wheat and rice grains.J Genet 93, 279-292. |
| [10] | Fulton DC, Edwards A, Pilling E, Robinson HL, Fahy B, Seale R, Kato L, Donald AM, Geigenberger P, Martin C, Smith AM (2002). Role of granule-bound starch synthase in determination of amylopectin structure and starch granule morphology in potato.J Biol Chem 277, 10834-10841. |
| [11] | Guzman C, Alvarez JB (2015). Wheat waxy proteins: polymorphism, molecular characterization and effects on starch properties.Theor Appl Genet 9, 1049-1060. |
| [12] | Hirose T, Hashida Y, Aoki N, Okamura M, Yonekura M, Ohto C, Terao T, Ohsugi R (2014). Analysis of gene- disruption mutants of a sucrose phosphate synthase gene in rice,OsSPS1, shows the importance of sucrose synthesis in pollen germination. Plant Sci 225, 102-106. |
| [13] | Hoai TT, Matsusaka H, Toyosawa Y, Suu TD, Satoh H, Kumamaru T (2014). Influence of single-nucleotide poly- morphisms in the gene encoding granule-bound starch synthase I on amylose content in Vietnamese rice cultivars.Breed Sci 64, 142-148. |
| [14] | Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y (2010). Starch biosynthesis in cereal endosperm.Plant Physiol Biochem 48, 383-392. |
| [15] | Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007). Clustal W and Clustal X version 2.0.Bioinformatics 23, 2947-2948. |
| [16] | Miao H, Sun P, Liu W, Xu B, Jin Z (2014). Identification of genes encoding granule-bound starch synthase involved in amylose metabolism in banana fruit.PLoS One 9, e88077. |
| [17] | Ohdan T, Francisco PB, Sawada TJ, Hirose T, Terao T, Satoh H, Nakamura Y (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice.J Exp Bot 56, 3229-3244. |
| [18] | Orzechowski S (2008). Starch metabolism in leaves.Acta Biochim Pol 55, 435-445. |
| [19] | Patron NJ, Smith AM, Fahy BF, Hylton CM, Naldrett MJ, Rossnagel BG, Denyer K (2002). The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5'-non-coding region.Plant Physiol 130, 190-198. |
| [20] | Tsai CY (1974). The function of the waxy locus in starch synthesis in maize endosperm.Biochem Genet 11, 83-96. |
| [21] | Vrinten PL, Nakamura T (2000). Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues.Plant Physiol 122, 255-264. |
| [22] | Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009). Jalview Version 2—a multiple sequence alignment editor and analysis workbench.Bioinformatics 25, 1189-1191. |
| [23] | Yan HB, Pan XX, Jiang HW, Wu GJ (2009). Comparison of the starch synthesis genes between maize and rice: copies, chromosome location and expression divergence.Theor Appl Genet 119, 815-825. |
| [24] | Zhu L, Gu M, Meng X, Cheung SC, Yu H, Huang J, Sun Y, Shi Y, Liu Q (2012). High-amylose rice improves indices of animal health in normal and diabetic rats.Plant Biotechnol J 10, 353-362. |
/
| 〈 |
|
〉 |