

花青素转录因子调控机制及代谢工程研究进展
收稿日期: 2018-01-15
录用日期: 2018-11-05
网络出版日期: 2018-12-10
基金资助
国家自然科学基金(31670305, No.31570306)
Recent Advances in the Regulation Mechanism of Transcription Factors and Metabolic Engineering of Anthocyanins
Received date: 2018-01-15
Accepted date: 2018-11-05
Online published: 2018-12-10
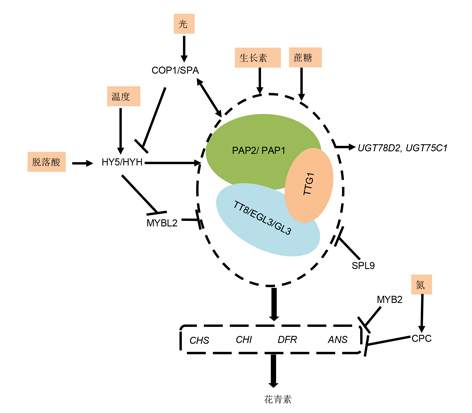
花青素是广泛存在于植物中的一类重要的类黄酮化合物, 在植物生长发育和人类营养保健方面具有重要价值。花青素的生物合成途径已经解析得比较清楚, 但花青素的代谢调控网络还在不断完善。调控花青素生物合成的转录因子主要包括MYB、bHLH和WD40三大类, 这些转录因子通过激活或抑制CHS、ANS和DFR等花青素途径关键结构基因的表达水平, 进而决定花青素积累的部位与水平。该文结合国内外花青素生物合成与转录调控方面的研究进展, 简要介绍了花青素的生物合成途径, 归纳总结了模式植物中花青素代谢调控的分子机理, 尤其是MYB、bHLH和WD40三类主要转录因子的调控机理, 以及这些转录因子在观赏植物和水果等经济作物花青素代谢工程中的应用。该文将为系统阐明花青素的转录调控机制和利用代谢工程改良花青素的相关研究提供有益参考。
宋雪薇,魏解冰,狄少康,庞永珍 . 花青素转录因子调控机制及代谢工程研究进展[J]. 植物学报, 2019 , 54(1) : 133 -156 . DOI: 10.11983/CBB18016
Anthocyanins are among the most important flavonoid compounds widely present in plants. Anthocyanins play significant roles in plant growth and development as well as human nutrition and health care. The anthocyanin biosynthetic pathway has been widely documented, and the anthocyanin metabolic regulation network is being constantly improved. The transcription factors that regulate anthocyanin biosynthesis mainly include three classes: MYB, bHLH and WD40 proteins. The proteins regulate the accumulation, location and levels of anthocyanins by activating or suppressing the expression of key structural genes, including CHS, ANS and DFR. This review briefly introduces the anthocyanin biosynthetic pathway and summarizes the molecular mechanism of transcriptional regulation based on recent progress. It mainly focuses on the molecular mechanism of MYB, bHLH and WD40 transcription factors in the regulation of anthocyanins in model plants. In addition, it summarizes the use of these transcription factors in anthocyanin metabolic engineering in ornamental plants and fruit crops. This review will provide valuable references for the in-depth investigation of transcriptional regulation and improving anthocyanins by metabolic engineering.

| 1 | 胡可, 韩科厅, 戴思兰 ( 2010). 环境因子调控植物花青素苷合成及呈色的机理. 植物学报 45, 307-317. |
| 2 | 朱丽, 钱前 ( 2017). 突破复杂性状多基因转化技术壁垒, 首创胚乳花青素高积累的水稻新种质. 植物学报 52, 539-542. |
| 3 | 祝志欣, 鲁迎青 ( 2016). 花青素代谢途径与植物颜色变异. 植物学报 51, 107-119. |
| 4 | Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi- Shinozaki K ( 2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63-78. |
| 5 | Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O'Connell AP ( 2001). The strawberry FaMYB1 transcription factor suppresses anthocyaninand flavonol accumulation in transgenic tobacco. Plant J 28, 319-332. |
| 6 | Albert NW, Davies KM, Lewis DH, Zhang HB, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE ( 2014). A conserved network of trans- criptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962-980. |
| 7 | Albert NW, Lewis DH, Zhang HB, Schwinn KE, Jameson PE, Davies KM ( 2011). Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning.Plant J 65, 771-784. |
| 8 | Almeida J, Carpenter R, Robbins TP, Martin C, Coen ES ( 1989). Genetic interactions underlying flower color pat- terns in Antirrhinum majus.Genes Dev 3, 1758-1767. |
| 9 | An JP, Li HH, Song LQ, Su L, Liu X, You CX, Wang XF, Hao YJ ( 2016). The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol Biochem 108, 24-31. |
| 10 | An JP, Qu FJ, Yao JF, Wang XN, You CX, Wang XF, Hao YJ ( 2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in ap- ple. Hortic Res 4, 17056. |
| 11 | An XH, Tian Y, Chen KQ, Liu XJ, Liu DD, Xie XB, Cheng CG, Cong PH, Hao YJ ( 2014). MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosyn- thesis of anthocyanin and proanthocyanidin in apples.Plant Cell Physiol 56, 650-662. |
| 12 | An XH, Tian Y, Chen KQ, Wang XF, Hao YJ ( 2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J Plant Physiol 169, 710-717. |
| 13 | Ang LH, Deng XW ( 1994). Regulatory hierarchy of photo- morphogenic loci: allele-specific and light-dependent inte- raction between the HY5 and COP1 loci.Plant Cell 6, 613-628. |
| 14 | Azuma A, Kobayashi S, Mitani N, Shiraishi M, Yamada M, Ueno T, Kono A, Yakushiji H, Koshita Y ( 2008). Geno- mic and genetic analysis of Myb-related genes that regu- late anthocyanin biosynthesis in grape berry skin.Theor Appl Genet 117, 1009-1019. |
| 15 | Bai YH, Pattanaik S, Patra B, Werkman JR, Xie CH, Yuan L ( 2011). Flavonoid-related basic helix-loop-helix regu- lators, NtAn1a and NtAn1b of tobacco have originated from two ancestors and are functionally active. Planta 234, 363-375. |
| 16 | Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T ( 2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48, 958-970. |
| 17 | Banerjee A, Roychoudhury A ( 2017). Abscisic-acid-depen- dent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254, 3-16. |
| 18 | Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L ( 2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana.Plant J 39, 366-380. |
| 19 | Ben-Simhon Z, Judeinstein S, Nadler-Hassar T, Trainin T, Bar-Ya’akov I, Borochov-Neori H, Holland D ( 2011). A pomegranate ( Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pome- granate fruit development.Planta 234, 865-881. |
| 20 | Boase MR, Lewis DH, Davies KM, Marshall GB, Patel D, Schwinn KE, Deroles SC ( 2010). Isolation and antisense suppression of flavonoid 3',5'-hydroxylase modifies flower pigments and colour in cyclamen. BMC Plant Biol 10, 107. |
| 21 | Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C ( 2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383-2393. |
| 22 | Broeckling BE, Watson RA, Steinwand B, Bush DR ( 2016). Intronic sequence regulates sugar-dependent ex- pression of Arabidopsis thaliana Production of Antho- cyanin Pigment-1/MYB75.PLoS One 11, e0156673. |
| 23 | Brueggemann J, Weisshaar B, Sagasser M ( 2010). A WD40-repeat gene from Malus × domestica is a functional homologue of Arabidopsis thaliana TRANSPARENT TES- TA GLABRA 1.Plant Cell Rep 29, 285-294. |
| 24 | Brugliera F, Tao GQ, Tems U, Kalc G, Mouradova E, Price K, Stevenson K, Nakamura N, Stacey I, Katsumoto Y, Tanaka Y, Mason JG ( 2013). Violet/blue chrysanthem ums—metabolic engineering of the anthocyanin biosyn- thetic pathway results in novel petal colors. Plant Cell Physiol 54, 1696-1710. |
| 25 | Butelli E, Licciardello C, Zhang Y, Liu JJ, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C ( 2012). Retrotransposons control fruit-specific, cold-dependent accu- mulation of anthocyanins in blood oranges. Plant Cell 24, 1242-1255. |
| 26 | Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, Martin C ( 2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26, 1301-1308. |
| 27 | Cabrita L, Fossen T, Andersen ?M ( 2000). Colour and stability of the six common anthocyanidin 3-glucosides in aqueous solutions. Food Chem 68, 101-107. |
| 28 | Carey CC, Strahle JT, Selinger DA, Chandler VL ( 2004). Mutations in the pale aleurone color 1 regulatory gene of the Zea mays anthocyanin pathway have distinct pheno- types relative to the functionally similar TRANSPARENT TESTA GLABRA 1 gene in Arabidopsis thaliana.Plant Cell 16, 450-464. |
| 29 | Cavallini E, Matus JT, Finezzo L, Zenoni S, Loyola R, Guzzo F, Schlechter R, Ageorges A, Arce-Johnson P, Tornielli GB ( 2015). The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol 167, 1448-1470. |
| 30 | Chagné D, Wang KL, Espley RV, Volz RK, How NM, Rouse S, Brendolise C, Carlisle CM, Kumar S, De Silva N, Micheletti D, McGhie T, Crowhurst RN, Storey RD, Velasco R, Hellens RP, Gardiner SE, Allan AC ( 2013). An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161, 225-239. |
| 31 | Chandler VL, Radicella JP, Robbins TP, Chen J, Turks D ( 1989). Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R geno- mic sequences.Plant Cell 1, 1175-1183. |
| 32 | Chen KL, Liu HL, Lou Q, Liu YL ( 2017). Corrigendum: ectopic expression of the grape hyacinth (Muscari armeniacum) R2R3-MYB transcription factor gene, MaAN2, induces anthocyanin accumulation in tobacco. Front Plant Sci 8, 1722. |
| 33 | Chiou CY, Yeh KW ( 2008). Differential expression of MYB gene ( OgMYB1) determines color patterning in floral tissue of Oncidium Gower Ramsey.Plant Mol Biol 66, 379-388. |
| 34 | Chiu LW, Li L ( 2012). Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosyn- thesis in purple cauliflower. Planta 236, 1153-1164. |
| 35 | Chiu LW, Zhou XJ, Burke S, Wu XL, Prior RL, Li L ( 2010). The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol 154, 1470-1480. |
| 36 | Cone KC, Burr FA, Burr B ( 1986). Molecular analysis of the maize anthocyanin regulatory locus C1.Proc Natl Acad Sci USA 83, 9631-9635. |
| 37 | Cone KC, Cocciolone SM, Burr FA, Burr B ( 1993 a). Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant.Plant Cell 5, 1795-1805. |
| 38 | Cone KC, Cocciolone SM, Moehlenkamp CA, Weber T, Drummond BJ, Tagliani LA, Bowen BA, Perrot GH (1993b). Role of the regulatory gene pl in the photocontrol of maize anthocyanin pigmentation.Plant Cell 5, 1807-1816. |
| 39 | Consonni G, Viotti A, Dellaporta SL, Tonelli C ( 1992). cDNA nucleotide sequence of Sn, a regulatory gene in maize.Nucleic Acids Res 20, 373. |
| 40 | Costantini L, Malacarne G, Lorenzi S, Troggio M, Mattivi F, Moser C, Grando MS ( 2015). New candidate genes for the fine regulation of the colour of grapes. J Exp Bot 66, 4427-4440. |
| 41 | Cultrone A, Cotroneo PS, Recupero GR ( 2010). Cloning and molecular characterization of R2R3-MYB and bHLH- MYC transcription factors from Citrus sinensis.Tree Genet Genom 6, 101-112. |
| 42 | Cutanda-Perez MC, Ageorges A, Gomez C, Vialet S, Terrier N, Romieu C, Torregrosa L ( 2009). Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport.Plant Mol Biol 69, 633-648. |
| 43 | de Vetten N, Quattrocchio F, Mol J, Koes R ( 1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals.Genes Dev 11, 1422-1434. |
| 44 | Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Mérillon JM, Hamdi S ( 2006). Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 149, 499-511. |
| 45 | Deluc L, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon JM, Robinson SP, Barrieu F ( 2008). The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol 147, 2041-2053. |
| 46 | Dixon RA, Sumner LW ( 2003). Legume natural products: understanding and manipulating complex pathways for human and animal health. Plant Physiol 131, 878-885. |
| 47 | Dong W, Niu LL, Gu JT, Gao F ( 2014). Isolation of a WD40-repeat gene regulating anthocyanin biosynthesis in storage roots of purple-fleshed sweet potato. Acta Physiol Plant 36, 1123-1132. |
| 48 | Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L ( 2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana.Plant J 55, 940-953. |
| 49 | Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L ( 2010). MYB transcription factors in Arabi- dopsis. Trends Plant Sci 15, 573-581. |
| 50 | Elomaa P, Uimari A, Mehto M, Albert VA, Laitinen RA, Teeri TH ( 2003). Activation of anthocyanin biosynthesis in Gerbera hybrida(Asteraceae) suggests conserved protein- protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol 133, 1831-1842. |
| 51 | Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, Allan AC ( 2009). Multiple repeats of a promoter segment causes transcription factor autoregu- lation in red apples. Plant Cell 21, 168-183. |
| 52 | Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC ( 2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49, 414-427. |
| 53 | Fan XP, Fan BH, Wang YX, Yang WC ( 2016). Anthocyanin accumulation enhanced in Lc-transgenic cotton under light and increased resistance to bollworm.Plant Biotechnol Rep 10, 1-11. |
| 54 | Farrell N, Norris G, Lee SG, Chun OK, Blesso CN ( 2015). Anthocyanin-rich black elderberry extract improves mar- kers of HDL function and reduces aortic cholesterol in hyperlipidemic mice. Food Funct 6, 1278-1287. |
| 55 | Feng SQ, Wang YL, Yang S, Xu YT, Chen XS ( 2010). Anthocyanin biosynthesis in pears is regulated by a R2R3- MYB transcription factor PyMYB10. Planta 232, 245-255. |
| 56 | Feyissa DN, L?vdal T, Olsen KM, Slimestad R, Lillo C ( 2009). The endogenous GL3, but not EGL3 gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves.Planta 230, 747-754. |
| 57 | Fraser LG, Seal AG, Montefiori M, McGhie TK, Tsang GK, Datson PM, Hilario E, Marsh HE, Dunn JK, Hellens RP, Davies KM, McNeilage MA, De Silva HN, Allan AC ( 2013). An R2R3 MYB transcription factor determines red petal colour in an Actinidia(kiwifruit) hybrid population. BMC Genomics 14, 28. |
| 58 | Fukusaki Ei, Kawasaki K, Kajiyama SI, An CI, Suzuki K, Tanaka Y, Kobayashi A ( 2004). Flower color modulations of Torenia hybrida by downregulation of chalcone syn- thase genes with RNA interference.J Biotechnol 111, 229-240. |
| 59 | Gao JJ, Shen XF, Zhang Z, Peng RH, Xiong AS, Xu J, Zhu B, Zheng JL, Yao QH ( 2011). The MYB transcription factor MdMYB6 suppresses anthocyanin biosynthesis in transgenic Arabidopsis. Plant Cell Tissue Organ Cult 106, 235-242. |
| 60 | Gatica-Arias A, Farag M, Stanke M, Matou?ek J, Wess- johann L, Weber G ( 2012). Flavonoid production in transgenic hop ( Humulus lupulus L.) altered by PAP1/ MYB75 from Arabidopsis thaliana L.Plant Cell Rep 31, 111-119. |
| 61 | Geekiyanage S, Takase T, Ogura Y, Kiyosue T ( 2007). Anthocyanin production by over-expression of grape transcription factor gene VlmybA2 in transgenic tobacco and Arabidopsis.Plant Biotechnol Rep 1, 11-18. |
| 62 | Gibalová A, Steinbachová L, Hafidh S, Bláhová V, Gadiou Z, Michailidis C, M?ller K, Pleskot R, Dup?áková N, Honys D ( 2017). Characterization of pollen-expressed bZIP protein interactions and the role of ATbZIP18 in the male gametophyte. Plant Reprod 30, 1-17. |
| 63 | Gong ZZ, Yamazaki M, Saito K ( 1999). A light-inducible Myb-like gene that is specifically expressed in red Perilla frutescens and presumably acts as a determining factor of the anthocyanin forma.Mol Gen Genet 262, 65-72. |
| 64 | Gonzalez A, Zhao MZ, Leavitt JM, Lloyd AM ( 2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53, 814-827. |
| 65 | Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW ( 2011). Negative regulation of anthocyanin biosynthesis in Ara- bidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512-1522. |
| 66 | He XZ, Li Y, Lawson D, Xie DY ( 2017). Metabolic engineer- ing of anthocyanins in dark tobacco varieties. Physiol Plant 159, 2-12. |
| 67 | Hichri I, Heppel SC, Pillet J, Léon C, Czemmel S, Delrot S, Lauvergeat V, Bogs J ( 2010). The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol Plant 3, 509-523. |
| 68 | Holton TA, Cornish EC ( 1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7, 1071-1083. |
| 69 | Hu QN, Yang L, Liu SD, Zhou LM, Wang XT, Wang W, Cai L, Wu XJ, Chang Y, Wang SC ( 2016). A repressor motif-containing poplar R3 MYB-like transcription factor regulates epidermal cell fate determination and antho- cyanin biosynthesis in Arabidopsis. J Plant Biol 59, 525-535. |
| 70 | Huang J, Wang MM, Bao YM, Sun SJ, Pan LJ, Zhang HS ( 2008). SRWD: a novel WD40 protein subfamily regulated by salt stress in rice ( Oryza sativa L.).Gene 424, 71-79. |
| 71 | Huang WJ, Lv HY, Wang Y ( 2017). Functional characteri- zation of a novel R2R3-MYB transcription factor modula- ting the flavonoid biosynthetic pathway from Epimedium sagittatum.Front Plant Sci 8, 1274. |
| 72 | Huang WJ, Sun W, Lv HY, Luo M, Zeng SH, Pattanaik S, Yuan L, Wang Y (2013a). A R2R3-MYB transcription factor from Epimedium sagittatum regulates the flavonoid biosynthetic pathway.PLoS One 8, e70778. |
| 73 | Huang YJ, Song S, Allan AC, Liu XF, Yin XR, Xu CJ, Chen KS ( 2013 b). Differential activation of anthocyanin biosyn- thesis in Arabidopsis and tobacco over-expressing an R2R3 MYB from Chinese bayberry. Plant Cell Tissue Organ Cult 113, 491-499. |
| 74 | Hughes NM, Neufeld HS, Burkey KO ( 2005). Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata.New Phytol 168, 575-587. |
| 75 | Isaak CK, Petkau JC, Blewett H, Karmin O, Siow YL ( 2017). Lingonberry anthocyanins protect cardiac cells from oxidative-stress-induced apoptosis. Can J Physiol Pharmacol 95, 904-910. |
| 76 | Jeong CY, Kim JH, Lee WJ, Jin JY, Kim J, Hong SW, Lee H ( 2018). AtMyb56 regulates anthocyanin levels via the modulation of AtGPT2 expression in response to sucrose in Arabidopsis.Mol Cells 41, 351-361. |
| 77 | Jo YN, Jin DE, Jeong JH, Kim HJ, Kim DO, Heo HJ ( 2015). Effect of anthocyanins from rabbit-eye blueberry ( Vacci- nium virgatum) on cognitive function in mice under trime- thyltin-induced neurotoxicity.Food Sci Biotechnol 24, 1077-1085. |
| 78 | Jun JH, Liu CG, Xiao XR, Dixon RA ( 2015). The transcrip- tional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula.Plant Cell 27, 2860-2879. |
| 79 | Jung CS, Griffiths HM, De Jong DM, Cheng SP, Bodis M, Kim TS, De Jong WS ( 2009). The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin.Theor Appl Genet 120, 45-57. |
| 80 | Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M, Nakamura N, Yonekura-Saka- kibara K, Togami J, Pigeaire A ( 2007). Engineering of the rose flavonoid biosynthetic pathway successfully gene- rated blue-hued flowers accumulating delphinidin. Plant Cell Physiol 48, 1589-1600. |
| 81 | Kobayashi S, Goto-Yamamoto N, Hirochika H ( 2004). Retrotransposon-induced mutations in grape skin color. Science 304, 982. |
| 82 | Kobayashi S, Ishimaru M, Hiraoka K, Honda C ( 2002). Myb-related genes of the Kyoho grape ( Vitis labruscana) regulate anthocyanin biosynthesis.Planta 215, 924-933. |
| 83 | Lai B, Du LN, Liu R, Hu B, Su WB, Qin YH, Zhao JT, Wang HC, Hu GB ( 2016). Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinen- sis during anthocyanin accumulation.Front Plant Sci 7, 166. |
| 84 | Lai B, Li XJ, Hu B, Qin YH, Huang XM, Wang HC, Hu GB ( 2014). LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes, tissues, developmental phases and ABA and light stimuli in Litchi chinensis.PLoS One 9, e86293. |
| 85 | Laitinen RAE, Ainasoja M, Broholm SK, Teeri TH, Elomaa P ( 2008). Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida.J Exp Bot 59, 3691-3703. |
| 86 | Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongp- rasit W, Zhao HY, Lee I, Deng XW ( 2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731-749. |
| 87 | Lee WJ, Jeong CY, Kwon J, Van Kien V, Lee D, Hong SW, Lee H ( 2016). Drastic anthocyanin increase in response to PAP1 overexpression in fls1 knockout mutant confers en- hanced osmotic stress tolerance in Arabidopsis thaliana.Plant Cell Rep 35, 2369-2379. |
| 88 | Li CH, Qiu J, Yang GS, Huang SR, Yin JM ( 2016 a). Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum(Hort.). Plant Cell Rep 35, 2151-2165. |
| 89 | Li N, Zong Y, Liu BL, Chen WJ, Zhang B ( 2017). TaMYB3, encoding a functional MYB transcriptor, isolated from the purple pericarp of Triticum aestivum.Cereal Res Commun 45, 369-380. |
| 90 | Li PH, Chen BB, Zhang GY, Chen LX, Dong Q, Wen JQ, Mysore KS, Zhao J ( 2016b). Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8.New Phytol 210, 905-921. |
| 91 | Li W, Wang B, Wang M, Chen M, Yin JM, Kaleri GM, Zhang RJ, Zuo TN, You X, Yang Q ( 2014). Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation.J Integr Plant Biol 56, 364-372. |
| 92 | Lim SH, Song JH, Kim DH, Kim JK, Lee JY, Kim YM, Ha SH ( 2016). Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1.Plant Cell Rep 35, 641-653. |
| 93 | Liu XF, Xiang LL, Yin XR, Grierson D, Li F, Chen KS ( 2015). The identification of a MYB transcription factor controlling anthocyanin biosynthesis regulation in Chry- santhemum flowers.Sci Hortic 194, 278-285. |
| 94 | Liu XF, Yin XR, Allan AC, Lin-Wang K, Shi YN, Huang YJ, Ferguson IB, Xu CJ, Chen KS ( 2013). The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosyn- thetic genes in tobacco and Chinese bayberry(Myrica rubra) during anthocyanin biosynthesis. Plant Cell Tissue Organ Cult 115, 285-298. |
| 95 | Liu Z, Shi MZ, Xie DY ( 2014). Regulation of anthocyanin biosynthesis in Arabidopsis thaliana red pap1-D cells metabolically programmed by auxins.Planta 239, 765-781. |
| 96 | Ludwig SR, Habera LF, Dellaporta SL, Wessler SR ( 1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc- homology region.Proc Natl Acad Sci USA 86, 7092-7096. |
| 97 | Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, Rubio V, Uhrig JF, Hülskamp M, Hoecker U ( 2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J 74, 638-651. |
| 98 | Mano H, Ogasawara F, Sato K, Higo H, Minobe Y ( 2007). Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol 143, 1252-1268. |
| 99 | Martin CA, Prescott A, Mackay S, Bartlett J, Vrijlandt E ( 1991). Control of anthocyanin biosynthesis in flowers of Antirrhinum majus.Plant J 1, 37-49. |
| 100 | Mathews H, Clendennen SK, Caldwell CG, Liu XL, Connors K, Matheis N, Schuster DK, Menasco D, Wagoner W, Lightner J, Wagner D ( 2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15, 1689-1703. |
| 101 | Matsui K, Umemura Y, Ohme-Takagi M ( 2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55, 954-967. |
| 102 | Matus JT, Poupin MJ, Ca?ón P, Bordeu E, Alcalde JA, Arce-Johnson P ( 2010). Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine(Vitis vinifera L.). Plant Mol Biol 72, 607-620. |
| 103 | Medina-Puche L, Cumplido-Laso G, Amil-Ruiz F, Hoff- mann T, Ring L, Rodríguez-Franco A, Caballero JL, Schwab W, Mu?oz-Blanco J, Blanco-Portales R ( 2014). MYB10 plays a major role in the regulation of flavonoid/ phenylpropanoid metabolism during ripening of Fragaria× ananassa fruits.J Exp Bot 65, 401-417. |
| 104 | Meyer P, Heidmann I, Forkmann G, Saedler H ( 1987). A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature 330, 677-678. |
| 105 | Miller JC, Chezem WR, Clay NK ( 2016). Ternary WD40 repeat-containing protein complexes: evolution, compo- sition and roles in plant immunity. Front Plant Sci 6, 1108. |
| 106 | Miller R, Owens SJ, R?rslett B ( 2011). Plants and colour: flowers and pollination. Opt Laser Technol 43, 282-294. |
| 107 | Mishra AK, Puranik S, Prasad M ( 2012). Structure and regulatory networks of WD40 protein in plants. J Plant Biochem Biotechnol 21, 32-39. |
| 108 | Mitsunami T, Nishihara M, Galis I, Alamgir KM, Hojo Y, Fujita K, Sasaki N, Nemoto K, Sawasaki T, Arimura GI ( 2014). Overexpression of the PAP1 transcription factor reveals a complex regulation of flavonoid and phenyl- propanoid metabolism in Nicotiana tabacum plants at- tacked by Spodoptera litura.PLoS One 9, e108849. |
| 109 | Morgenstern B, Atchley WR ( 1999). Evolution of bHLH transcription factors: modular evolution by domain shuf- fling? Mol Biol Evol 16, 1654-1663. |
| 110 | Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S ( 2006). Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory.Plant Cell Physiol 47, 457-470. |
| 111 | Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, Michael AJ, Tohge T, Yamazaki M, Saito K ( 2014). Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77, 367-379. |
| 112 | Nakatsuka A, Yamagishi M, Nakano M, Tasaki K, Kobayashi N ( 2009). Light-induced expression of basic helix-loop-helix genes involved in anthocyanin biosyn- thesis in flowers and leaves of asiatic hybrid lily. Sci Hortic 121, 84-91. |
| 113 | Nakatsuka T, Haruta KS, Pitaksutheepong C, Abe Y, Kakizaki Y, Yamamoto K, Shimada N, Yamamura S, Nishihara M ( 2008). Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Phy- siol 49, 1818-1829. |
| 114 | Nemie-Feyissa D, Olafsdottir SM, Heidari B, Lillo C ( 2014). Nitrogen depletion and small R3-MYB transcription factors affecting anthocyanin accumulation in Arabidopsis leaves. Phytochemistry 98, 34-40. |
| 115 | Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L ( 2000). The TT8 gene encodes a basic helix- loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863-1878. |
| 116 | Nguyen NH, Jeong CY, Kang GH, Yoo SD, Hong SW, Lee H ( 2015). MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis.Plant J 84, 1192-1205. |
| 117 | Niu SS, Xu CJ, Zhang WS, Zhang B, Li X, Lin-Wang K, Ferguson IB, Allan AC, Chen KS ( 2010). Coordinated regulation of anthocyanin biosynthesis in Chinese bay- berry ( Myrica rubra) fruit by a R2R3 MYB transcription factor.Planta 231, 887-899. |
| 118 | Noda N, Yoshioka S, Kishimoto S, Nakayama M, Douzono M, Tanaka Y, Aida R ( 2017). Generation of blue chrysan- themums by anthocyanin B-ring hydroxylation and glu- cosylation and its coloration mechanism. Sci Adv 3, e160-2785. |
| 119 | Ohno S, Hosokawa M, Hoshino A, Kitamura Y, Morita Y, Park KI, Nakashima A, Deguchi A, Tatsuzawa F Doi M, Iida S, Yazawa S, (2011).A bHLH transcription factor,DvIVS, is involved in regulation of anthocyanin synthesis indahlia(Dahlia variabilis). J Exp Bot 62, 5105-5116. |
| 120 | Onkokesung N, Reichelt M, van Doorn A, Schuurink RC, van Loon JJ, Dicke M ( 2014). Modulation of flavonoid metabolites in Arabidopsis thaliana through overex- pression of the MYB75 transcription factor: role of kaem- pferol-3,7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae.J Exp Bot 65, 2203-2217. |
| 121 | Palapol Y, Ketsa S, Lin-Wang K, Ferguson IB, Allan AC ( 2009). A MYB transcription factor regulates anthocyanin biosynthesis in mangosteen ( Garcinia mangostana L.) fruit during ripening.Planta 229, 1323-1334. |
| 122 | Pang YZ, Wenger JP, Saathoff K, Peel GJ, Wen JQ, Huhman D, Allen SN, Tang YH, Cheng XF, Tadege M, Ratet P, Mysore KS, Sumner LW, Marks MD, Dixon RA ( 2009). A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proan- thocyanidin biosynthesis but not for trichome development.Plant Physiol 151, 1114-1129. |
| 123 | Park JS, Kim JB, Cho KJ, Cheon CI, Sung MK, Choung GM, Roh KH ( 2008). Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce ( Lactuca sativa).Plant Cell Rep 27, 985-994. |
| 124 | Park K, Ishikawa N, Morita Y, Choi J, Hoshino A, Iida S ( 2007). A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosyn- thesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation.Plant J 49, 641-654. |
| 125 | Passeri V, Martens S, Carvalho E, Bianchet C, Damiani F, Paolocci F ( 2017). The R2R3MYB VvMYBPA1 from grape reprograms the phenylpropanoid pathway in tobacco flowers.Planta 246, 185-199. |
| 126 | Pattanaik S, Kong Q, Zaitlin D, Werkman JR, Xie CH, Patra B, Yuan L ( 2010). Isolation and functional char- acterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 231, 1061-1076. |
| 127 | Payne CT, Zhang F, Lloyd AM ( 2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Ge- netics 156, 1349-1362. |
| 128 | Payyavula RS, Singh RK, Navarre DA ( 2013). Trans- cription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J Exp Bot 64, 5115-5131. |
| 129 | Paz-Ares J, Ghosal D, Wienand U, Peterson P, Saedler H ( 1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators.EMBO J 6, 3553-3558. |
| 130 | Peel GJ, Pang YZ, Modolo LV, Dixon RA ( 2009). The LAP1 MYB transcription factor orchestrates anthocyanidin bio- synthesis and glycosylation in Medicago.Plant J 59, 136-149. |
| 131 | Peiffer DS, Wang LS, Zimmerman NP, Ransom BWS, Carmella SG, Kuo CT, Chen JH, Oshima K, Huang YW, Hecht SS, Stoner GD ( 2016). Dietary consumption of black raspberries or their anthocyanin constituents alters innate immune cell trafficking in esophageal cancer. Cancer Immunol Res 4, 72-82. |
| 132 | Pelletier MK, Murrell JR, Shirley BW ( 1997). Charac- terization of flavonol synthase and leucoanthocyanidin dioxygenase genes in arabidopsis: further evidence for differential regulation of "Early" and "Late" genes. Plant Physiol 113, 1437-1445. |
| 133 | Pérez-Díaz JR, Pérez-Díaz J, Madrid-Espinoza J, González- Villanueva E, Moreno Y, Ruiz-Lara S ( 2016). New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol Biol 90, 63-76. |
| 134 | Pierantoni L, Dondini L, De Franceschi P, Musacchi S, Winkel BSJ, Sansavini S ( 2010). Mapping of an anthocyanin-regulating MYB transcription factor and its expression in red and green pear, Pyrus communis.Plant Physiol Biochem 48, 1020-1026. |
| 135 | Pooma W, Gersos C, Grotewold E ( 2002). Transposon insertions in the promoter of the Zea mays a1 gene differentially affect transcription by the Myb factors P and C1.Genetics 161, 793-801. |
| 136 | Qiu J, Gao FH, Shen GA, Li CH, Han XY, Zhao Q, Zhao DX, Hua XJ, Pang YZ ( 2013). Metabolic engineering of the phenylpropanoid pathway enhances the antioxidant capacity of Saussurea involucrata.PLoS One 8, e70665. |
| 137 | Qiu J, Sun SQ, Luo SQ, Zhang JC, Xiao XZ, Zhang LQ, Wang F, Liu SZ ( 2014). Arabidopsis AtPAP1 transcription factor induces anthocyanin production in transgenic Taraxacum brevicorniculatum.Plant Cell Rep 33, 669-680. |
| 138 | Qiu ZK, Wang XX, Gao JC, Guo YM, Huang ZJ, Du YC ( 2016). The tomato Hoffman’s Anthocyaninless gene encodes a bHLH transcription factor involved in ant- hocyanin biosynthesis that is developmentally regulated and induced by low temperatures.PLoS One 11, e015-1067. |
| 139 | Quattrocchio F, Wing J, Van der Woude K, Souer E, de Vetten N, Mol J, Koes R ( 1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color.Plant Cell 11, 1433-1444. |
| 140 | Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE ( 1993). Regulatory genes controlling anthocyanin pigmen- tation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5, 1497-1512. |
| 141 | Quattrocchio F, Wing JF, Van der Woude K, Mol JNM, Koes R ( 1998). Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are cau- sed by divergent evolution of target anthocyanin genes. Plant J 13, 475-488. |
| 142 | Ramsay NA, Walker AR, Mooney M, Gray JC ( 2003). Two basic-helix-loop-helix genes ( MYC-146 and GL3) from Arabidopsis can activate anthocyanin biosynthesis in a white-flowered Matthiola incana mutant.Plant Mol Biol 52, 679-688. |
| 143 | Ravaglia D, Espley RV, Henry-Kirk RA, Andreotti C, Ziosi V, Hellens RP, Costa G, Allan AC ( 2013). Transcriptional regulation of flavonoid biosynthesis in nectarine ( Prunus persica) by a set of R2R3 MYB transcription factors.BMC Plant Biol 13, 68. |
| 144 | Rommens CM, Richael CM, Yan H, Navarre DA, Ye JS, Krucker M, Swords K ( 2008). Engineered native path- ways for high kaempferol and caffeoylquinate production in potato. Plant Biotechnol J 6, 870-886. |
| 145 | Salvatierra A, Pimentel P, Moya-León MA, Herrera R ( 2013). Increased accumulation of anthocyanins in Fra- garia chiloensis fruits by transient suppression of FcMYB1 gene.Phytochemistry 90, 25-36. |
| 146 | Schwinn K, Venail J, Shang YJ, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C ( 2006). A small family of MYB-regulatory genes controls floral pig- mentation intensity and patterning in the genus Antir- rhinum.Plant Cell 18, 831-851. |
| 147 | Schwinn KE, Ngo H, Kenel F, Brummell DA, Albert NW, McCallum JA, Pither-Joyce M, Crowhurst RN, Eady C, Davies KM ( 2016). The onion ( Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis.Front Plant Sci 7, 1865. |
| 148 | Seeram NP, Nair MG ( 2002). Inhibition of lipid peroxidation and structure—activity-related studies of the dietary con- stituents anthocyanins, anthocyanidins, and catechins. J Agric Food Chem 50, 5308-5312. |
| 149 | Shao L, Shu Z, Sun SL, Peng CL, Wang XJ, Lin ZF ( 2007). Antioxidation of anthocyanins in photosynthesis under high temperature stress. J Integr Plant Biol 49, 1341-1351. |
| 150 | Shen XJ, Zhao K, Liu LL, Zhang KC, Yuan HZ, Liao X, Wang Q, Guo XW, Li F, Li TH ( 2014). A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng ( Prunus avium L.).Plant Cell Physiol 55, 862-880. |
| 151 | Shi MZ, Xie DY ( 2011). Engineering of red cells of Arabidop- sis thaliana and comparative genome-wide gene expres- sion analysis of red cells versus wild-type cells.Planta 233, 787-805. |
| 152 | Shih PH, Yeh CT, Yen GC ( 2007). Anthocyanins induce the activation of phase II enzymes through the antioxidant response element pathway against oxidative stress-indu- ced apoptosis. J Agric Food Chem 55, 9427-9435. |
| 153 | Shim SH, Kim JM, Choi CY, Kim CY, Park KH ( 2012). Ginkgo biloba extract and bilberry anthocyanins improve visual function in patients with normal tension glaucoma. J Med Food 15, 818-823. |
| 154 | Shimizu Y, Maeda K, Kato M, Shimomura K ( 2011). Co-expression of GbMYB1 and GbMYC1 induces antho- cyanin accumulation in roots of cultured Gynura bicolor DC.plantlet on methyl jasmonate treatment. Plant Physiol Biochem 49, 159-167. |
| 155 | Shin DH, Choi M, Kim K, Bang G, Cho M, Choi SB, Choi G, Park YI ( 2013). HY5 regulates anthocyanin biosyn- thesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett 587, 1543-1547. |
| 156 | Shin DH, Choi MG, Kang CS, Park CS, Choi SB, Park YI ( 2016). A wheat R2R3-MYB protein PURPLE PLANT 1 (TaPL1) functions as a positive regulator of anthocyanin biosynthesis. Biochem Biophys Res Commun 469, 686-691. |
| 157 | Shin J, Park E, Choi G ( 2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49, 981-994. |
| 158 | Sompornpailin K, Makita Y, Yamazaki M, Saito K ( 2002). A WD-repeat-containing putative regulatory protein in antho- cyanin biosynthesis in Perilla frutescens.Plant Mol Biol 50, 485-495. |
| 159 | Song SS, Qi TC, Fan M, Zhang X, Gao H, Huang H, Wu DW, Guo HW, Xie DX ( 2013). The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant de- fense and development. PLoS Genet 9, e1003653. |
| 160 | Song SS, Qi TC, Huang H, Ren QC, Wu DW, Chang CQ, Peng W, Liu YL, Peng JR, Xie DX ( 2011). The jasmonate-ZIM domain proteins interact with the R2R3- MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000-1013. |
| 161 | Spelt C, Quattrocchio F, Mol JNM, Koes R ( 2000). antho- cyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural antho- cyanin genes. Plant Cell 12, 1619-1632. |
| 162 | Starkevi? P, Pauk?tyt? J, Kazanavi?iūt? V, Denkovskien? E, Stanys V, Bendokas V, ?ik?nianas T, Ra?anskien? A, Ra?anskas R ( 2015). Expression and anthocyanin biosynthesis-modulating potential of sweet cherry ( Prunus avium L.) MYB10 and bHLH genes.PLoS One 10, e012-6991. |
| 163 | Sun L, Fan XC, Zhang Y, Jiang JF, Sun HS, Liu CH ( 2016). Transcriptome analysis of genes involved in anthocyanins biosynthesis and transport in berries of black and white spine grapes ( Vitis davidii).Hereditas 153, 17. |
| 164 | Takahashi R, Benitez ER, Oyoo ME, Khan NA, Komatsu S ( 2011). Nonsense mutation of an MYB transcription factor is associated with purple-blue flower color in soybean. J Hered 102, 458-463. |
| 165 | Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR ( 2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples.Plant Physiol 142, 1216-1232. |
| 166 | Tao Y, Chen T, Yang GQ, Peng GH, Yan ZJ, Huang YF ( 2016). Anthocyanin can arrest the cone photoreceptor degeneration and act as a novel treatment for retinitis pigmentosa. Int J Ophthalmol 9, 153-158. |
| 167 | Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima Ji, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M ( 2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42, 218-235. |
| 168 | Toledo-Ortiz G, Huq E, Quail PH ( 2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749-1770. |
| 169 | Tonelli C, Consonni G, Dolfini SF, Dellaporta SL, Viotti A, Gavazzi G ( 1991). Genetic and molecular analysis of Sn, a light-inducible, tissue specific regulatory gene in maize.Mol Gen Genet 225, 401-410. |
| 170 | Urao T, Yamaguchi-Shinozaki K, Mitsukawa N, Shibata D, Shinozaki K ( 1996). Molecular cloning and characteri- zation of a gene that encodes a MYC-related protein in Arabidopsis. Plant Mol Biol 32, 571-576. |
| 171 | Vimolmangkang S, Han YP, Wei GC, Korban SS ( 2013). An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower deve- lopment. BMC Plant Biol 13, 176. |
| 172 | Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC ( 1999). The TRANSPARENT TESTA GLABRA 1 locus, which regulates trichome differentiation and antho- cyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein.Plant Cell 11, 1337-1350. |
| 173 | Wang CL, Lu GQ, Hao YQ, Guo HM, Guo Y, Zhao J, Cheng HM ( 2017). ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in trans- genic cotton. Planta 246, 453-469. |
| 174 | Wang KL, Bolitho K, Grafton K, Kortstee A, Karun airetnam S, McGhie TK, Espley RV, HellensRP, Allan AC ( 2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10, 50. |
| 175 | Wang YL, Wang YQ, Song ZQ, Zhang HY ( 2016). Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis.Mol Plant 9, 1395-1405. |
| 176 | Wang ZG, Meng D, Wang AD, Li TL, Jiang SL, Cong PH, Li TZ ( 2013). The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear.Plant Physiol 162, 885-896. |
| 177 | Wei JY, Wu HJ, Zhang HQ, Li F, Chen SR, Hou BH, Shi YH, Zhao LJ, Duan HJ ( 2018). Anthocyanins inhibit high glucose-induced renal tubular cell apoptosis caused by oxidative stress in db/db mice. Int J Mol Med 41, 1608-1618. |
| 178 | Winkel-Shirley B ( 2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and bio- technology. Plant Physiol 126, 485-493. |
| 179 | Xiang LL, Liu XF, Li X, Yin XR, Grierson D, Li F, Chen KS ( 2015). A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in chrysanthemums ( Chrysanthemum morifolium Ramat.).PLoS One 10, e0143892. |
| 180 | Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ ( 2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35, 1884-1897. |
| 181 | Xie XB, Zhao J, Hao YJ, Fang CB, Wang Y ( 2017). The ectopic expression of apple MYB1 and bHLH3 differentially activates anthocyanin biosynthesis in tobacco.Plant Cell Tiss Organ Cul 131, 183-194. |
| 182 | Xu F, Ning Y, Zhang W, Liao Y, Li L, Cheng H, Cheng S ( 2014 a). An R2R3-MYB transcription factor as a negative regulator of the flavonoid biosynthesis pathway in Ginkgo biloba.Funct Integr Genomics 14, 177-189. |
| 183 | Xu W, Zhang N, Jiao Y, Li R, Xiao D, Wang Z ( 2014 b). The grapevine basic helix-loop-helix (bHLH) transcription factor positively modulates CBF-pathway and confers tolerance to cold-stress in Arabidopsis. Mol Biol Rep 41, 5329-5342. |
| 184 | Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K ( 2010). Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily.Plant Cell Physiol 51, 463-474. |
| 185 | Yamagishi M, Toda S, Tasaki K ( 2014). The novel allele of the LhMYB12 gene is involved in splatter-type spot for- mation on the flower tepals of Asiatic hybrid lilies(Lilium spp.). New Phytol 201, 1009-1020. |
| 186 | Yamazaki M, Makita Y, Springob K, Saito K ( 2003). Regulatory mechanisms for anthocyanin biosynthesis in chemotypes of Perilla frutescens var.crispa. Biochem Eng J 3, 191-197. |
| 187 | Yan YJ, Chemler J, Huang LX, Martens S, Koffas MAG ( 2005). Metabolic engineering of anthocyanin biosynthe- sis in Escherichia coli.Appl Environ Microbiol 71, 3617-3623. |
| 188 | Yao GF, Ming ML, Allan AC, Gu C, Li LT, Wu X, Wang RZ, Chang YJ, Qi KJ, Zhang SL, Wu J ( 2017 a). Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis.Plant J 92, 437-451. |
| 189 | Yao PF, Zhao HX, Luo XP, Gao F, Li Cl, Yao HP, Chen H, Park SU, Wu Q ( 2017 b). Fagopyrum tataricum FtWD40 functions as a positive regulator of anthocyanin biosyn- thesis in transgenic tobacco. J Plant Growth Regul 36, 755-765. |
| 190 | Yuan YX, Chiu LW, Li L ( 2009). Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta 230, 1141-1153. |
| 191 | Zhang B, Hu ZL, Zhang YJ, Li YL, Zhou S, Chen GP ( 2012). A putative functional MYB transcription factor induced by low temperature regulates anthocyanin bio- synthesis in purple kale ( Brassica Oleracea var.acephala f. tricolor). Plant Cell Rep 31, 281-289. |
| 192 | Zhang F, Gonzalez A, Zhao MZ, Payne CT, Lloyd A ( 2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859-4869. |
| 193 | Zhang LL, Wang Y, Sun M, Wang J, Kawabata S, Li YH ( 2014 a). BrMYB4, a suppressor of genes for phenyl- propanoid and anthocyanin biosynthesis, is down-regul- ated by UV-B but not by pigment-inducing sunlight in turnip cv. Tsuda. Plant Cell Physiol 55, 2092-2101. |
| 194 | Zhang LY, Bai MY, Wu JX, Zhu JY, Wang H, Zhang ZG, Wang WF, Sun Y, Zhao J, Sun XH, Yang HJ, Xu YY, Kim SH, Fujioka S, Lin WH, Chong K, Lu TG, Wang ZY ( 2009). Antagonistic HLH/bHLH transcription factors me- diate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21, 3767-3780. |
| 195 | Zhang Q, Hao RJ, Xu ZD, Yang WR, Wang J, Cheng TR, Pan HT, Zhang QX ( 2017). Isolation and functional characterization of a R2R3-MYB regulator of Prunus mu- me anthocyanin biosynthetic pathway.Plant Cell Tissue Organ Cult 131, 417-429. |
| 196 | Zhang WW, Xu F, Cheng SY, Liao YL ( 2018). Charac- terization and functional analysis of a MYB gene ( Gb- MYBFL) related to ?avonoid accumulation in Ginkgo biloba.Genes Genom 40, 49-61. |
| 197 | Zhang Y, Butelli E, Martin C ( 2014 b). Engineering antho- cyanin biosynthesis in plants. Curr Opin Plant Biol 19, 81-90. |
| 198 | Zhao DQ, Tao J ( 2015). Recent advances on the deve- lopment and regulation of flower color in ornamental plants. Front Plant Sci 6, 261. |
| 199 | Zhou LL, Shi MZ, Xie DY ( 2012). Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-pro- grammed red cells of Arabidopsis thaliana.Planta 236, 825-837. |
| 200 | Zhu HF, Fitzsimmons K, Khandelwal A, Kranz RG ( 2009). CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant 2, 790-802. |
| 201 | Zhu QL, Yu SZ, Zeng DC, Liu HM, Wang HC, Yang ZF, Xie XR, Shen RX, Tan JT, Li HY, Zhao XC, Zhang QY, Chen YL, Guo JX, Chen LT, Liu YG ( 2017). Development of “Purple Endosperm Rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency trans- gene stacking system. Mol Plant 10, 918-929. |
| 202 | Zhu YN, Xia M, Yang Y, Liu FQ, Li ZX, Hao YT, Mi MT, Jin TR, Ling WH ( 2011). Purified anthocyanin supplemen- tation improves endothelial function via NO-cGMP acti- vation in hypercholesterolemic individuals. Clin Chem 57, 1524-1533. |
/
| 〈 |
|
〉 |