

Chinese Bulletin of Botany ›› 2015, Vol. 50 ›› Issue (1): 55-71.DOI: 10.3724/SP.J.1259.2015.00055
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Ying Bao1, 2, *, Changfeng Guo1, Shaohua Chen1, Mei Liu1
Received:2013-11-04
Accepted:2014-02-23
Online:2015-01-01
Published:2015-04-09
Contact:
Bao Ying
About author:? These authors contributed equally to this paper
Ying Bao, Changfeng Guo, Shaohua Chen, Mei Liu. Molecular Evolution of Chalcone Synthase Gene Superfamily in Plants[J]. Chinese Bulletin of Botany, 2015, 50(1): 55-71.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.chinbullbotany.com/EN/10.3724/SP.J.1259.2015.00055
| Species | Locus No. | Gene ID | Strand | Chromosome | Duplication pattern |
|---|---|---|---|---|---|
| Arabidopsis thaliana | 1 | AT1G02050 | - | 1 | |
| 2 | AT4G00040 | + | 4 | ||
| 3 | AT4G34850 | + | 4 | ||
| 4 | AT5G13930 | + | 5 | ||
| Medicago truncatula | 1 | MT1G097900 | + | 1 | Tandem |
| 2 | MT1G097910 | + | 1 | Tandem | |
| 3 | MT1G098140 | + | 1 | Tandem | |
| 4 | MT1G098150 | + | 1 | Tandem | |
| 5 | MT2G058470 | - | 2 | ||
| 6 | MT3G083910 | - | 3 | Tandem and block | |
| 7 | MT3G083920 | - | 3 | Tandem and block | |
| 8 | MT3G086260 | + | 3 | ||
| 9 | MT4G078730 | + | 4 | ||
| 10 | MT5G007720 | + | 5 | Tandem | |
| 11 | MT5G007730 | + | 5 | Tandem | |
| 12 | MT5G007740 | + | 5 | Tandem | |
| 13 | MT5G007760 | + | 5 | Tandem | |
| 14 | MT5G007770 | + | 5 | Tandem | |
| 15 | MT7G016700 | + | 7 | Tandem | |
| 16 | MT7G016720 | - | 7 | Tandem | |
| 17 | MT7G016780 | + | 7 | Tandem | |
| 18 | MT7G016800 | + | 7 | Tandem | |
| 19 | MT7G016820 | + | 7 | Tandem | |
| 20 | MT7G084300 | - | 7 | ||
| 21 | MT8G085200 | + | 8 | Block | |
| Populus trichocarpa | 1 | PT00G02200 | - | Scaffold_955 | |
| 2 | PT01G06410 | + | Scaffold_1 | Block | |
| 3 | PT01G14120 | + | Scaffold_1 | Tandem and block | |
| 4 | PT01G14130 | + | Scaffold_1 | Tandem and block | |
| 5 | PT02G14050 | - | Scaffold_2 | Block | |
| 6 | PT03G16580 | + | Scaffold_3 | Tandem and block | |
| 7 | PT03G16590 | + | Scaffold_3 | Tandem and block | |
| 8 | PT03G16600 | + | Scaffold_3 | Tandem and block | |
| 9 | PT04G16510 | + | Scaffold_4 | Block | |
| 10 | PT05G15150 | - | Scaffold_5 | Tandem | |
| 11 | PT09G12950 | + | Scaffold_9 | Block | |
| 12 | PT12G12680 | + | Scaffold_12 | Block | |
| 13 | PT14G05350 | - | Scaffold_14 | Block | |
| 14 | PT14G14010 | - | Scaffold_14 | ||
| Vitis vinifera | 1 | VV03G05390 | + | 3 | |
| 2 | VV05G00090 | + | 5 | ||
| 3 | VV10G06850 | + | 10 | Tandem | |
| 4 | VV10G06860 | + | 10 | Tandem | |
| 5 | VV14G13530 | + | 14 | ||
| 6 | VV15G00770 | - | 15 | ||
| 7 | VV16G00180 | - | 16 | Tandem | |
| 8 | VV16G00200 | - | 16 | Tandem | |
| Species | Locus No. | Gene ID | Strand | Chromosome | Duplication pattern |
| 9 | VV16G00220 | - | 16 | Tandem | |
| 10 | VV16G00240 | - | 16 | Tandem | |
| 11 | VV16G00260 | - | 16 | Tandem | |
| 12 | VV16G00280 | + | 16 | Tandem | |
| 13 | VV16G00290 | - | 16 | Tandem | |
| 14 | VV16G00310 | - | 16 | Tandem | |
| 15 | VV16G00320 | - | 16 | Tandem | |
| 16 | VV16G00330 | - | 16 | Tandem | |
| 17 | VV16G00340 | - | 16 | Tandem | |
| 18 | VV16G00350 | - | 16 | Tandem | |
| 19 | VV16G00360 | - | 16 | Tandem | |
| 20 | VV16G00370 | - | 16 | Tandem | |
| 21 | VV16G00380 | + | 16 | Tandem | |
| 22 | VV16G00390 | + | 16 | Tandem | |
| Oryza sativa | 1 | OS01G41834 | - | 1 | |
| 2 | OS04G01354 | - | 4 | ||
| 3 | OS04G23940 | - | 4 | ||
| 4 | OS05G12180 | - | 5 | Tandem | |
| 5 | OS05G12190 | - | 5 | Tandem | |
| 6 | OS05G12210 | - | 5 | Tandem | |
| 7 | OS05G12240 | - | 5 | Tandem | |
| 8 | OS05G41645 | + | 5 | ||
| 9 | OS07G11440 | - | 7 | ||
| 10 | OS07G17010 | + | 7 | ||
| 11 | OS07G22850 | - | 7 | ||
| 12 | OS07G31750 | - | 7 | Tandem | |
| 13 | OS07G31770 | - | 7 | Tandem | |
| 14 | OS07G34140 | - | 7 | Tandem | |
| 15 | OS07G34190 | - | 7 | Tandem | |
| 16 | OS07G34260 | - | 7 | Tandem | |
| 17 | OS10G07040 | + | 10 | ||
| 18 | OS10G07616 | + | 10 | ||
| 19 | OS10G08620 | + | 10 | Tandem | |
| 20 | OS10G08670 | + | 10 | Tandem | |
| 21 | OS10G08710 | + | 10 | Tandem | |
| 22 | OS10G09860 | - | 10 | ||
| 23 | OS10G34360 | + | 10 | ||
| 24 | OS11G32540 | - | 11 | Tandem | |
| 25 | OS11G32580 | - | 11 | Tandem | |
| 26 | OS11G32610 | + | 11 | Tandem | |
| 27 | OS11G32620 | - | 11 | Tandem | |
| 28 | OS11G32650 | - | 11 | Tandem | |
| 29 | OS11G35930 | + | 11 | ||
| 30 | OS12G07690 | - | 12 | ||
| Zea mays | 1 | ZM01G41780 | - | 1 | Block |
| 2 | ZM02G42820 | - | 2 | Tandem and block | |
| 3 | ZM02G42850 | - | 2 | Tandem and block | |
| 4 | ZM02G45550 | + | 2 | ||
| Species | Locus No. | Gene ID | Strand | Chromosome | Duplication pattern |
| 5 | ZM03G09860 | + | 3 | Tandem | |
| 6 | ZM03G09890 | - | 3 | Tandem | |
| 7 | ZM03G09930 | - | 3 | Tandem | |
| 8 | ZM04G30630 | + | 4 | Tandem and block | |
| 9 | ZM04G30650 | + | 4 | Tandem and block | |
| 10 | ZM04G32440 | - | 4 | ||
| 11 | ZM05G09820 | + | 5 | Block | |
| 12 | ZM05G24850 | + | 5 | ||
| 13 | ZM07G22450 | - | 7 | Block | |
| Physcomitrella patens | 1 | PP00001G00030 | - | Scaffold_1 | |
| 2 | PP00020G01360 | - | Scaffold_20 | ||
| 3 | PP00022G00030 | + | Scaffold_22 | ||
| 4 | PP00025G01920 | + | Scaffold_25 | Tandem | |
| 5 | PP00025G01930 | - | Scaffold_25 | Tandem | |
| 6 | PP00034G01040 | + | Scaffold_34 | ||
| 7 | PP00038G00030 | - | Scaffold_38 | ||
| 8 | PP00039G01610 | - | Scaffold_39 | ||
| 9 | PP00061G00280 | - | Scaffold_61 | ||
| 10 | PP00076G01020 | + | Scaffold_76 | ||
| 11 | PP00228G00140 | - | Scaffold_228 | Tandem | |
| 12 | PP00292G00010 | - | Scaffold_292 | ||
| 13 | PP00303G00060 | - | Scaffold_303 | Tandem | |
| 14 | PP00303G00070 | + | Scaffold_303 | Tandem | |
| 15 | PP00304G00340 | - | Scaffold_304 | ||
| 16 | PP00365G00100 | - | Scaffold_365 | Tandem | |
| 17 | PP00365G00120 | + | Scaffold_365 | Tandem | |
| 18 | PP00425G00060 | - | Scaffold_425 | Tandem | |
| 19 | PP00426G00290 | - | Scaffold_426 | ||
| 20 | PP00463G00060 | - | Scaffold_463 | Tandem | |
| 21 | PP00463G00070 | + | Scaffold_463 | Tandem | |
| 22 | PP00463G00100 | - | Scaffold_463 | Tandem | |
| 23 | PP00500G00030 | - | Scaffold_500 | ||
| Selaginella moellendorffii | 1 | SM00001G06800 | - | Scaffold_1 | |
| 2 | SM00017G03910 | - | Scaffold_17 | ||
| 3 | SM00007G01560 | - | Scaffold_7 | ||
| 4 | SM00068G00810 | - | Scaffold_68 | ||
| Ostreococcus lucimarinus | - | - | - | - | - |
| O. tauri | - | - | - | - | - |
| Micromonas sp. RCC299 | - | - | - | - | - |
| Volvox carteri | - | - | - | - | - |
| Chlamydomonas reinhardtii | - | - | - | - | - |
Table 1 Genes in CHS superfamily identified from 13 plants genomes by BLAST searching
| Species | Locus No. | Gene ID | Strand | Chromosome | Duplication pattern |
|---|---|---|---|---|---|
| Arabidopsis thaliana | 1 | AT1G02050 | - | 1 | |
| 2 | AT4G00040 | + | 4 | ||
| 3 | AT4G34850 | + | 4 | ||
| 4 | AT5G13930 | + | 5 | ||
| Medicago truncatula | 1 | MT1G097900 | + | 1 | Tandem |
| 2 | MT1G097910 | + | 1 | Tandem | |
| 3 | MT1G098140 | + | 1 | Tandem | |
| 4 | MT1G098150 | + | 1 | Tandem | |
| 5 | MT2G058470 | - | 2 | ||
| 6 | MT3G083910 | - | 3 | Tandem and block | |
| 7 | MT3G083920 | - | 3 | Tandem and block | |
| 8 | MT3G086260 | + | 3 | ||
| 9 | MT4G078730 | + | 4 | ||
| 10 | MT5G007720 | + | 5 | Tandem | |
| 11 | MT5G007730 | + | 5 | Tandem | |
| 12 | MT5G007740 | + | 5 | Tandem | |
| 13 | MT5G007760 | + | 5 | Tandem | |
| 14 | MT5G007770 | + | 5 | Tandem | |
| 15 | MT7G016700 | + | 7 | Tandem | |
| 16 | MT7G016720 | - | 7 | Tandem | |
| 17 | MT7G016780 | + | 7 | Tandem | |
| 18 | MT7G016800 | + | 7 | Tandem | |
| 19 | MT7G016820 | + | 7 | Tandem | |
| 20 | MT7G084300 | - | 7 | ||
| 21 | MT8G085200 | + | 8 | Block | |
| Populus trichocarpa | 1 | PT00G02200 | - | Scaffold_955 | |
| 2 | PT01G06410 | + | Scaffold_1 | Block | |
| 3 | PT01G14120 | + | Scaffold_1 | Tandem and block | |
| 4 | PT01G14130 | + | Scaffold_1 | Tandem and block | |
| 5 | PT02G14050 | - | Scaffold_2 | Block | |
| 6 | PT03G16580 | + | Scaffold_3 | Tandem and block | |
| 7 | PT03G16590 | + | Scaffold_3 | Tandem and block | |
| 8 | PT03G16600 | + | Scaffold_3 | Tandem and block | |
| 9 | PT04G16510 | + | Scaffold_4 | Block | |
| 10 | PT05G15150 | - | Scaffold_5 | Tandem | |
| 11 | PT09G12950 | + | Scaffold_9 | Block | |
| 12 | PT12G12680 | + | Scaffold_12 | Block | |
| 13 | PT14G05350 | - | Scaffold_14 | Block | |
| 14 | PT14G14010 | - | Scaffold_14 | ||
| Vitis vinifera | 1 | VV03G05390 | + | 3 | |
| 2 | VV05G00090 | + | 5 | ||
| 3 | VV10G06850 | + | 10 | Tandem | |
| 4 | VV10G06860 | + | 10 | Tandem | |
| 5 | VV14G13530 | + | 14 | ||
| 6 | VV15G00770 | - | 15 | ||
| 7 | VV16G00180 | - | 16 | Tandem | |
| 8 | VV16G00200 | - | 16 | Tandem | |
| Species | Locus No. | Gene ID | Strand | Chromosome | Duplication pattern |
| 9 | VV16G00220 | - | 16 | Tandem | |
| 10 | VV16G00240 | - | 16 | Tandem | |
| 11 | VV16G00260 | - | 16 | Tandem | |
| 12 | VV16G00280 | + | 16 | Tandem | |
| 13 | VV16G00290 | - | 16 | Tandem | |
| 14 | VV16G00310 | - | 16 | Tandem | |
| 15 | VV16G00320 | - | 16 | Tandem | |
| 16 | VV16G00330 | - | 16 | Tandem | |
| 17 | VV16G00340 | - | 16 | Tandem | |
| 18 | VV16G00350 | - | 16 | Tandem | |
| 19 | VV16G00360 | - | 16 | Tandem | |
| 20 | VV16G00370 | - | 16 | Tandem | |
| 21 | VV16G00380 | + | 16 | Tandem | |
| 22 | VV16G00390 | + | 16 | Tandem | |
| Oryza sativa | 1 | OS01G41834 | - | 1 | |
| 2 | OS04G01354 | - | 4 | ||
| 3 | OS04G23940 | - | 4 | ||
| 4 | OS05G12180 | - | 5 | Tandem | |
| 5 | OS05G12190 | - | 5 | Tandem | |
| 6 | OS05G12210 | - | 5 | Tandem | |
| 7 | OS05G12240 | - | 5 | Tandem | |
| 8 | OS05G41645 | + | 5 | ||
| 9 | OS07G11440 | - | 7 | ||
| 10 | OS07G17010 | + | 7 | ||
| 11 | OS07G22850 | - | 7 | ||
| 12 | OS07G31750 | - | 7 | Tandem | |
| 13 | OS07G31770 | - | 7 | Tandem | |
| 14 | OS07G34140 | - | 7 | Tandem | |
| 15 | OS07G34190 | - | 7 | Tandem | |
| 16 | OS07G34260 | - | 7 | Tandem | |
| 17 | OS10G07040 | + | 10 | ||
| 18 | OS10G07616 | + | 10 | ||
| 19 | OS10G08620 | + | 10 | Tandem | |
| 20 | OS10G08670 | + | 10 | Tandem | |
| 21 | OS10G08710 | + | 10 | Tandem | |
| 22 | OS10G09860 | - | 10 | ||
| 23 | OS10G34360 | + | 10 | ||
| 24 | OS11G32540 | - | 11 | Tandem | |
| 25 | OS11G32580 | - | 11 | Tandem | |
| 26 | OS11G32610 | + | 11 | Tandem | |
| 27 | OS11G32620 | - | 11 | Tandem | |
| 28 | OS11G32650 | - | 11 | Tandem | |
| 29 | OS11G35930 | + | 11 | ||
| 30 | OS12G07690 | - | 12 | ||
| Zea mays | 1 | ZM01G41780 | - | 1 | Block |
| 2 | ZM02G42820 | - | 2 | Tandem and block | |
| 3 | ZM02G42850 | - | 2 | Tandem and block | |
| 4 | ZM02G45550 | + | 2 | ||
| Species | Locus No. | Gene ID | Strand | Chromosome | Duplication pattern |
| 5 | ZM03G09860 | + | 3 | Tandem | |
| 6 | ZM03G09890 | - | 3 | Tandem | |
| 7 | ZM03G09930 | - | 3 | Tandem | |
| 8 | ZM04G30630 | + | 4 | Tandem and block | |
| 9 | ZM04G30650 | + | 4 | Tandem and block | |
| 10 | ZM04G32440 | - | 4 | ||
| 11 | ZM05G09820 | + | 5 | Block | |
| 12 | ZM05G24850 | + | 5 | ||
| 13 | ZM07G22450 | - | 7 | Block | |
| Physcomitrella patens | 1 | PP00001G00030 | - | Scaffold_1 | |
| 2 | PP00020G01360 | - | Scaffold_20 | ||
| 3 | PP00022G00030 | + | Scaffold_22 | ||
| 4 | PP00025G01920 | + | Scaffold_25 | Tandem | |
| 5 | PP00025G01930 | - | Scaffold_25 | Tandem | |
| 6 | PP00034G01040 | + | Scaffold_34 | ||
| 7 | PP00038G00030 | - | Scaffold_38 | ||
| 8 | PP00039G01610 | - | Scaffold_39 | ||
| 9 | PP00061G00280 | - | Scaffold_61 | ||
| 10 | PP00076G01020 | + | Scaffold_76 | ||
| 11 | PP00228G00140 | - | Scaffold_228 | Tandem | |
| 12 | PP00292G00010 | - | Scaffold_292 | ||
| 13 | PP00303G00060 | - | Scaffold_303 | Tandem | |
| 14 | PP00303G00070 | + | Scaffold_303 | Tandem | |
| 15 | PP00304G00340 | - | Scaffold_304 | ||
| 16 | PP00365G00100 | - | Scaffold_365 | Tandem | |
| 17 | PP00365G00120 | + | Scaffold_365 | Tandem | |
| 18 | PP00425G00060 | - | Scaffold_425 | Tandem | |
| 19 | PP00426G00290 | - | Scaffold_426 | ||
| 20 | PP00463G00060 | - | Scaffold_463 | Tandem | |
| 21 | PP00463G00070 | + | Scaffold_463 | Tandem | |
| 22 | PP00463G00100 | - | Scaffold_463 | Tandem | |
| 23 | PP00500G00030 | - | Scaffold_500 | ||
| Selaginella moellendorffii | 1 | SM00001G06800 | - | Scaffold_1 | |
| 2 | SM00017G03910 | - | Scaffold_17 | ||
| 3 | SM00007G01560 | - | Scaffold_7 | ||
| 4 | SM00068G00810 | - | Scaffold_68 | ||
| Ostreococcus lucimarinus | - | - | - | - | - |
| O. tauri | - | - | - | - | - |
| Micromonas sp. RCC299 | - | - | - | - | - |
| Volvox carteri | - | - | - | - | - |
| Chlamydomonas reinhardtii | - | - | - | - | - |
| No. | Contig | Length (bp) | Score bits | Strand | Identities | E-value | Sequence information |
|---|---|---|---|---|---|---|---|
| 1 | Ctg7180044571087 | 2 448 | 1 564 | - | 1 134/1 301 (87%) | 0 | Complete |
| 2 | Ctg7180044837494 | 4 493 | 1 535 | - | 1 119/1 301 (86%) | 0 | Complete |
| 3 | Ctg7180046059388 | 2 761 | 1 736 | - | 1 185/1 328 (89%) | 0 | Complete |
| 4 | Ctg7180055825794 | 3 986 | 1 000; 241 | - - | 821/999 (82%); 180/211 (85%) | 0 7e-60 | Complete |
| 5 | Ctg7180056125030 | 4 720 | 1 476; 378 | - - | 976/1 077 (91%); 226/237 (95%) | 0 4e-101 | Complete |
| 6 | Ctg7180057362889 | 39 003 | 1 893 | - | 1 265/1 402 (90%) | 0 | Complete |
| 7 | Jtg7180055651041f_7180057409216f | 5 769 | 1 337; 309 | - - | 891/991 (90%); 215/244 (88%) | 0 2e-80 | Complete |
| 8 | Jtg7180057127125f_7180046406791r | 10 190 | 1 577 | - | 1 123/1 284 (87%) | 0 | Complete |
| 9 | Ctg7180057114010 | 6 665 | 1 342 | + | 1 077/1 298 (83%) | 0 | Complete |
| 10 | Ctg7180057423160 | 15 081 | 1 471 | + | 1 087/1 271 (86%) | 0 | Complete |
| 11 | Jtg7180046403543f_7180056907730f | 6 806 | 2 006 | + | 1 288/1 401 (92%) | 0 | Complete |
| 12 | Jtg7180043014796r_7180045416866r | 5 549 | 717; 149 | + + | 762/1 001 (76%); 141/180 (78%) | 0 3e-32 | Complete |
| 13 | Ctg7180057238333 | 10 113 | 1 712; 342 | - - | 966/977 (99%); 194/197 (98%) | 0 2e-90 | Complete |
| 14 | Ctg7180055304970 | 1 885 | 1 265 | - | 911/1 039 (88%) | 0 | Fragment |
| 15 | Ctg7180044078159 | 1 566 | 1 209 | - | 875/1 000 (88%) | 0 | Fragment |
| 16 | Ctg7180054149681 | 1 521 | 951 | + | 646/724 (89%) | 0 | Fragment |
| 17 | Ctg7180040742080 | 576 | 872 | - | 539/576 (94%) | 0 | Fragment |
| 18 | Ctg7180055755024 | 954 | 805 | + | 537/596 (90%) | 0 | Fragment |
| 19 | Ctg7180054149682 | 656 | 792 | + | 507/552 (92%) | 0 | Fragment |
| 20 | Ctg7180057090732 | 1 429 | 735 | - | 766/1 001 (77%) | 0 | Fragment |
| 21 | Ctg7180056804266 | 2 400 | 735 | + | 766/1 001 (77%) | 0 | Fragment |
| 22 | Ctg7180055304969 | 1 187 | 726 | - | 476/524 (91%) | 0 | Fragment |
| 23 | Deg7180050331267 | 530 | 704 | + | 474/530 (89%) | 0 | Fragment |
| 24 | Ctg7180047225082 | 2 089 | 704 | + | 756/998 (76%) | 0 | Fragment |
| 25 | Ctg7180057563133 | 1 376 | 675 | + | 753/1 002 (75%) | 0 | Fragment |
| 26 | Deg7180049910668 | 707 | 659 | - | 574/706 (81%) | 0 | Fragment |
| 27 | Ctg7180040473811 | 2 084 | 650 | + | 734/981 (75%) | 0 | Fragment |
| 28 | Deg7180050301638 | 1 145 | 623 | + | 510/616 (83%) | 6e-175 | Fragment |
| 29 | Ctg7180046407516 | 2 023 | 600 | + | 736/1 002 (73%) | 6e-168 | Fragment |
| 30 | Ctg7180056729621 | 1 548 | 583 | + | 654/871 (75%) | 5e-163 | Fragment |
| 31 | Ctg7180055812743 | 7 348 | 583 | - | 730/999 (73%) | 5e-163 | Fragment |
| 32 | Ctg7180056199763 | 7 255 | 574 | + | 722/987 (73%) | 3e-160 | Fragment |
| 33 | Ctg7180055845812 | 12 473 | 563 | - | 667/900 (74%) | 5e-157 | Fragment |
| 34 | Ctg7180053679462 | 1 148 | 554 | - | 659/891 (74%) | 2e-154 | Fragment |
| 35 | Ctg7180050291419 | 893 | 554 | + | 609/806 (76%) | 2e-154 | Fragment |
| 36 | Ctg7180040305101 | 946 | 553 | + | 623/833 (75%) | 8e-154 | Fragment |
| 37 | Ctg7180056889828 | 1 745 | 545 | + | 622/831 (75%) | 1e-151 | Fragment |
| 38 | Ctg7180040359262 | 801 | 544 | + | 536/691 (78%) | 4e-151 | Fragment |
| 39 | Ctg7180038459346 | 430 | 533 | - | 376/430 (87%) | 8e-148 | Fragment |
| 40 | Ctg7180038501054 | 559 | 522 | - | 453/555 (82%) | 1e-144 | Fragment |
| 41 | Ctg7180050814991 | 1 172 | 509 | + | 600/808 (74%) | 9e-141 | Fragment |
| 42 | Ctg7180057409908 | 1 097 | 508 | + | 343/384 (89%) | 3e-140 | Fragment |
| No. | Contig | Length (bp) | Score bits | Strand | Identities | E-value | Sequence information |
| 43 | Ctg7180056950914 | 6 228 | 1 357 | + | 1 092/1 318 (83%) | 0 | Nonsense mutation |
| 44 | Ctg7180056550739 | 18 693 | 1 342 | - | 1 076/1 293 (83%) | 0 | Nonsense mutation |
| 45 | Jtg7180056551170f_7180056551171f | 9 504 | 1 198 | - | 1 060/1 326 (80%) | 0 | Nonsense mutation |
| 46 | Ctg7180039529527 | 1 759 | 677 | + | 754/1 001 (75%) | 0 | Nonsense mutation |
| 47 | Jtg7180057668388f_7180057668389f | 5 289 | 661 | + | 734/975 (75%) | 0 | Nonsense mutation |
| 48 | Ctg7180044573223 | 1 796 | 650 | - | 747/1 001 (75%) | 0 | Nonsense mutation |
| 49 | Ctg7180056441624 | 5 881 | 600 | + | 739/1 002 (74%) | 6e-168 | Nonsense mutation |
| 50 | Jtg7180056265080f_7180046063071r | 7 390 | 513 | + | 714/992 (72%) | 7e-142 | Nonsense mutation |
| 51 | Jtg7180057409910f_7180057409911f | 20 052 | 1 393 | + | 1 080/1 281 (84%) | 0 | Frame shift |
Table 2 Contigs contained genes of CHS superfamily in Pinus taeda genome
| No. | Contig | Length (bp) | Score bits | Strand | Identities | E-value | Sequence information |
|---|---|---|---|---|---|---|---|
| 1 | Ctg7180044571087 | 2 448 | 1 564 | - | 1 134/1 301 (87%) | 0 | Complete |
| 2 | Ctg7180044837494 | 4 493 | 1 535 | - | 1 119/1 301 (86%) | 0 | Complete |
| 3 | Ctg7180046059388 | 2 761 | 1 736 | - | 1 185/1 328 (89%) | 0 | Complete |
| 4 | Ctg7180055825794 | 3 986 | 1 000; 241 | - - | 821/999 (82%); 180/211 (85%) | 0 7e-60 | Complete |
| 5 | Ctg7180056125030 | 4 720 | 1 476; 378 | - - | 976/1 077 (91%); 226/237 (95%) | 0 4e-101 | Complete |
| 6 | Ctg7180057362889 | 39 003 | 1 893 | - | 1 265/1 402 (90%) | 0 | Complete |
| 7 | Jtg7180055651041f_7180057409216f | 5 769 | 1 337; 309 | - - | 891/991 (90%); 215/244 (88%) | 0 2e-80 | Complete |
| 8 | Jtg7180057127125f_7180046406791r | 10 190 | 1 577 | - | 1 123/1 284 (87%) | 0 | Complete |
| 9 | Ctg7180057114010 | 6 665 | 1 342 | + | 1 077/1 298 (83%) | 0 | Complete |
| 10 | Ctg7180057423160 | 15 081 | 1 471 | + | 1 087/1 271 (86%) | 0 | Complete |
| 11 | Jtg7180046403543f_7180056907730f | 6 806 | 2 006 | + | 1 288/1 401 (92%) | 0 | Complete |
| 12 | Jtg7180043014796r_7180045416866r | 5 549 | 717; 149 | + + | 762/1 001 (76%); 141/180 (78%) | 0 3e-32 | Complete |
| 13 | Ctg7180057238333 | 10 113 | 1 712; 342 | - - | 966/977 (99%); 194/197 (98%) | 0 2e-90 | Complete |
| 14 | Ctg7180055304970 | 1 885 | 1 265 | - | 911/1 039 (88%) | 0 | Fragment |
| 15 | Ctg7180044078159 | 1 566 | 1 209 | - | 875/1 000 (88%) | 0 | Fragment |
| 16 | Ctg7180054149681 | 1 521 | 951 | + | 646/724 (89%) | 0 | Fragment |
| 17 | Ctg7180040742080 | 576 | 872 | - | 539/576 (94%) | 0 | Fragment |
| 18 | Ctg7180055755024 | 954 | 805 | + | 537/596 (90%) | 0 | Fragment |
| 19 | Ctg7180054149682 | 656 | 792 | + | 507/552 (92%) | 0 | Fragment |
| 20 | Ctg7180057090732 | 1 429 | 735 | - | 766/1 001 (77%) | 0 | Fragment |
| 21 | Ctg7180056804266 | 2 400 | 735 | + | 766/1 001 (77%) | 0 | Fragment |
| 22 | Ctg7180055304969 | 1 187 | 726 | - | 476/524 (91%) | 0 | Fragment |
| 23 | Deg7180050331267 | 530 | 704 | + | 474/530 (89%) | 0 | Fragment |
| 24 | Ctg7180047225082 | 2 089 | 704 | + | 756/998 (76%) | 0 | Fragment |
| 25 | Ctg7180057563133 | 1 376 | 675 | + | 753/1 002 (75%) | 0 | Fragment |
| 26 | Deg7180049910668 | 707 | 659 | - | 574/706 (81%) | 0 | Fragment |
| 27 | Ctg7180040473811 | 2 084 | 650 | + | 734/981 (75%) | 0 | Fragment |
| 28 | Deg7180050301638 | 1 145 | 623 | + | 510/616 (83%) | 6e-175 | Fragment |
| 29 | Ctg7180046407516 | 2 023 | 600 | + | 736/1 002 (73%) | 6e-168 | Fragment |
| 30 | Ctg7180056729621 | 1 548 | 583 | + | 654/871 (75%) | 5e-163 | Fragment |
| 31 | Ctg7180055812743 | 7 348 | 583 | - | 730/999 (73%) | 5e-163 | Fragment |
| 32 | Ctg7180056199763 | 7 255 | 574 | + | 722/987 (73%) | 3e-160 | Fragment |
| 33 | Ctg7180055845812 | 12 473 | 563 | - | 667/900 (74%) | 5e-157 | Fragment |
| 34 | Ctg7180053679462 | 1 148 | 554 | - | 659/891 (74%) | 2e-154 | Fragment |
| 35 | Ctg7180050291419 | 893 | 554 | + | 609/806 (76%) | 2e-154 | Fragment |
| 36 | Ctg7180040305101 | 946 | 553 | + | 623/833 (75%) | 8e-154 | Fragment |
| 37 | Ctg7180056889828 | 1 745 | 545 | + | 622/831 (75%) | 1e-151 | Fragment |
| 38 | Ctg7180040359262 | 801 | 544 | + | 536/691 (78%) | 4e-151 | Fragment |
| 39 | Ctg7180038459346 | 430 | 533 | - | 376/430 (87%) | 8e-148 | Fragment |
| 40 | Ctg7180038501054 | 559 | 522 | - | 453/555 (82%) | 1e-144 | Fragment |
| 41 | Ctg7180050814991 | 1 172 | 509 | + | 600/808 (74%) | 9e-141 | Fragment |
| 42 | Ctg7180057409908 | 1 097 | 508 | + | 343/384 (89%) | 3e-140 | Fragment |
| No. | Contig | Length (bp) | Score bits | Strand | Identities | E-value | Sequence information |
| 43 | Ctg7180056950914 | 6 228 | 1 357 | + | 1 092/1 318 (83%) | 0 | Nonsense mutation |
| 44 | Ctg7180056550739 | 18 693 | 1 342 | - | 1 076/1 293 (83%) | 0 | Nonsense mutation |
| 45 | Jtg7180056551170f_7180056551171f | 9 504 | 1 198 | - | 1 060/1 326 (80%) | 0 | Nonsense mutation |
| 46 | Ctg7180039529527 | 1 759 | 677 | + | 754/1 001 (75%) | 0 | Nonsense mutation |
| 47 | Jtg7180057668388f_7180057668389f | 5 289 | 661 | + | 734/975 (75%) | 0 | Nonsense mutation |
| 48 | Ctg7180044573223 | 1 796 | 650 | - | 747/1 001 (75%) | 0 | Nonsense mutation |
| 49 | Ctg7180056441624 | 5 881 | 600 | + | 739/1 002 (74%) | 6e-168 | Nonsense mutation |
| 50 | Jtg7180056265080f_7180046063071r | 7 390 | 513 | + | 714/992 (72%) | 7e-142 | Nonsense mutation |
| 51 | Jtg7180057409910f_7180057409911f | 20 052 | 1 393 | + | 1 080/1 281 (84%) | 0 | Frame shift |
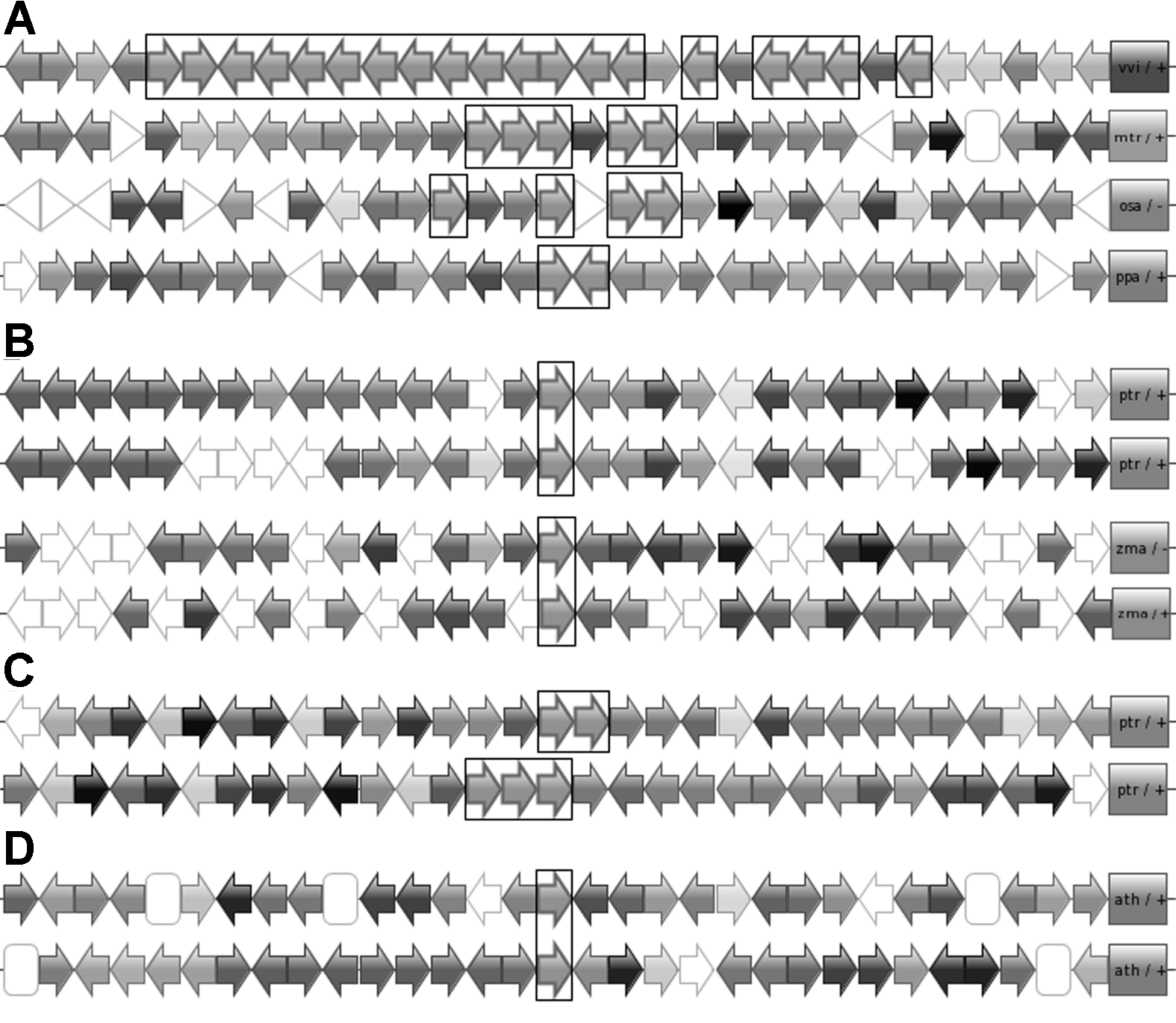
Figure 1 Three duplication patterns of genes occurred in CHS superfamily (A) Patterns of tandem duplication in Vitis vinifera, Medicago truncatula, Oryza sativa and Physcomitrella patens; (B) Patterns of block duplication between Populus trichocarpa and V. vinifera; (C) Pattern of block and tandem duplication in P. trichocarpa; (D) Pattern of possible transposition duplication in Arabidopsis thaliana. Arrows represent genes with direction in genomes; Homologues shows in same colors; Target genes in CHS superfamily are framed
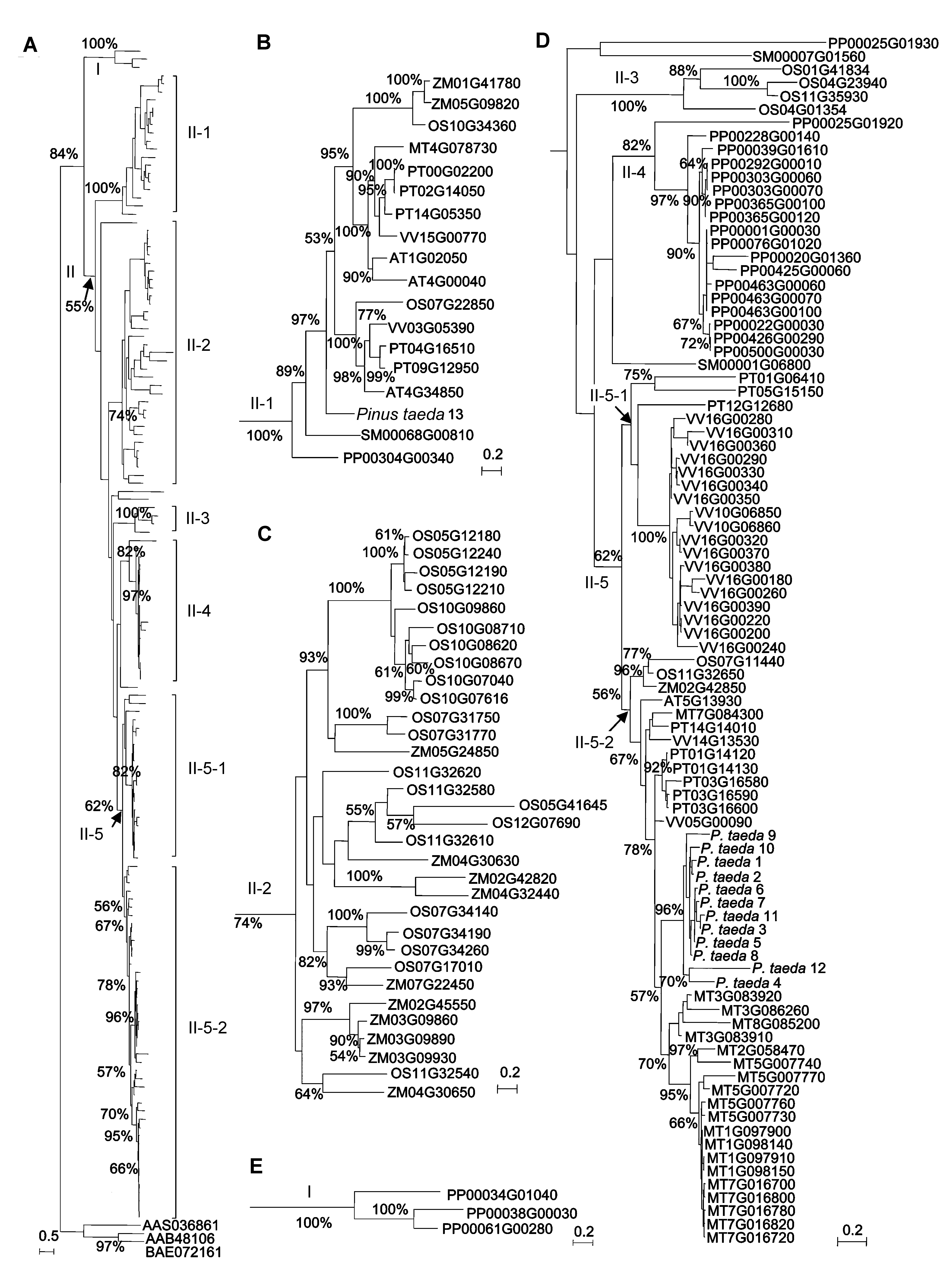
Figure 2 Phylogram resulting of the CHS superfamily from maximum likelihood analysis with an amino acid substitution model LG+G+F (A) Whole tree; (B)-(E) Clades in detail. Numbers beside the branches refer to the bootstrap values (>50%) based on 100 replications. Gene information is shown in Table 1 and 2.
| 1 | Abe I, Morita H (2010). Structure and function of the chalcone synthase superfamily of plant type III poly- ketide synthases. Nat Prod Rep 27, 809-838. |
| 2 | Austin MB, Noel JP (2003). The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep 20, 79-110. |
| 3 | Baerson SR, Schröder J, Cook D, Rimando AM, Pan ZQ, Dayan FE, Noonan BP, Duke SO (2010). Alkylresorcinol biosynthesis in plants: new insights from an ancient enzyme family? Plant Signal Behav 5, 1286-1289. |
| 4 | Bowers JE, Chapman BA, Rong JK, Paterson AH (2003). Unravelling angiosperm genome evolution by phylo- genetic analysis of chromosomal duplication events.Nature 422, 433-438. |
| 5 | Christensen AB, Gregersen PL, Schröder J, Collinge DB (1998). A chalcone synthase with an unusual substrate preference is expressed in barley leaves in response to UV light and pathogen attack. Plant Mol Biol 37, 849-857. |
| 6 | Cook D, Rimando AM, Clemente TE, Schröder J, Dayan FE, Nanayakkara NP, Pan ZQ, Noonan BP, Fishbein M, Abe I, Duke SO, Baerson SR (2010). Alkylresorcinol synthases expressed in Sorghum bicolor root hairs play an essential role in the biosynthesis of the allelopathic benzoquinone sorgoleone. Plant Cell 22, 867-887. |
| 7 | Criscuolo A (2011). MorePhyML: improving the phylo- genetic tree space exploration with PhyML 3.Mol Phylo- genet Evol 61, 944-948. |
| 8 | Darriba D, Taboada GL, Doallo R, Posada D (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164-1165. |
| 9 | Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW (2010). LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153, 937-955. |
| 10 | Durbin ML, Learn GH Jr, Huttley GA, Clegg MT (1995). Evolution of the chalcone synthase gene family in the genus Ipomoea. Proc Natl Acad Sci USA 92, 3338-3342. |
| 11 | Durbin ML, McCaig B, Clegg MT (2000). Molecular evolution of the chalcone synthase multigene family in the morning glory genome. Plant Mol Biol 42, 79-92. |
| 12 | Ferrer JL, Austin MB, Stewart C Jr, Noel JP (2008). Structure and function of enzymes involved in the bio- synthesis of phenylpropanoids.Plant Physiol Bioch 46, 356-370. |
| 13 | Fliegmann J, Schröder G, Schanz S, Britsch L, Schröder J (1992). Molecular analysis of chalcone and dihydro- pinosylvin synthase from Scots pine (Pinus sylvestris), and differential regulation of these and related enzyme activities in stressed plants. Plant Mol Biol 18, 489-503. |
| 14 | Franken P, Niesbach-Klösgen U, Weydemann U, Maré- chal-Drouard L, Saedler H, Wienand U (1991). The duplicated chalcone synthase genes C2 and Whp (white pollen) of Zea mays are independently regulated; evid- ence for translational control of Whp expression by the anthocyanin intensifying gene in. EMBO J 10, 2605-2612. |
| 15 | Freeling M (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60, 433-453. |
| 16 | Fukada-Tanaka S, Hoshino A, Hisatomi Y, Habu Y, Hasebe M, Iida S (1997). Identification of new chalcone synthase genes for flower pigmentation in the Japanese and common morning glories. Plant Cell Physiol 38, 754-758. |
| 17 | Funa N, Awakawa T, Horinouchi S (2007). Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa.J Biol Chem 282, 14476-14481. |
| 18 | Funa N, Ozawa H, Hirata A, Horinouchi S (2006). Phenolic lipid synthesis by type III polyketide synthases is essential for cyst formation in Azotobacter vinelandii. Proc Natl Acad Sci USA 103, 6356-6361. |
| 19 | Gross F, Luniak N, Perlova O, Gaitatzis N, Jenke- Kodama H, Gerth K, Gottschalk D, Dittmann E, Muller R (2006). Bacterial type III polyketide synthases: phy- logenetic analysis and potential for the production of novel secondary metabolites by heterologous expression in pseudomonads. Arch Microbiol 185, 28-38. |
| 20 | Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH (2008). Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environ- mental stimuli. Plant Physiol 148, 993-1003. |
| 21 | Hihara Y, Hara C, Uchimiya H (1996). Isolation and characterization of two cDNA clones for mRNAs that are abundantly expressed in immature anthers of rice (Oryza sativa L.). Plant Mol Biol 30, 1181-1193. |
| 22 | Hopwood DA, Sherman DH (1990). Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet 24, 37-66. |
| 23 | Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyère C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pè ME, Valle G, Mor- gante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quétier F, Wincker P (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463-467. |
| 24 | Jez JM, Ferrer JL, Bowman ME, Austin MB, Schröder J, Dixon RA, Noel JP (2001). Structure and mechanism of chalcone synthase-like polyketide synthases. J Ind Mi- crobiol Biot 27, 393-398. |
| 25 | Jiang C, Kim SY, Suh DY (2008). Divergent evolution of the thiolase superfamily and chalcone synthase family. Mol Phylogenet Evol 49, 691-701. |
| 26 | Jiang CG, Schommer CK, Kim SY, Suh DY (2006). Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens. Phytochemistry 67, 2531-2540. |
| 27 | Jiao YN, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang HY, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, de Pamphilis CW (2011). Ancestral polyploidy in seed plants and angio- sperms. Nature 473, 97-100. |
| 28 | Katsuyama Y, Matsuzawa M, Funa N, Horinouchi S (2007).In vitro synthesis of curcuminoids by type III polyketide synthase from Oryza sativa. J Biol Chem 282, 37702-37709. |
| 29 | Kim SS, Grienenberger E, Lallemand B, Colpitts CC, Kim SY, Souza Cde A, Geoffroy P, Heintz D, Krahn D, Kaiser M, Kombrink E, Heitz T, Suh DY, Legrand M, Douglas CJ (2010). LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydro- xyalkyl α-pyrone synthases required for pollen develop- ment and sporopollenin biosynthesis in Arabidopsis tha- liana. Plant Cell 22, 4045-4066. |
| 30 | Kodan A, Kuroda H, Sakai F (2002). A stilbene synthase from Japanese red pine (Pinus densiflora): implications for phytoalexin accumulation and down-regulation of flavonoid biosynthesis. Proc Natl Acad Sci USA 99, 3335-3339. |
| 31 | Koduri PK, Gordon GS, Barker EI, Colpitts CC, Ashton NW, Suh DY (2010). Genome-wide analysis of the chal- cone synthase superfamily genes of Physcomitrella patens. Plant Mol Biol 72, 247-263. |
| 32 | Koes RE, Spelt CE, van den Elzen PJ, Mol JNM (1989). Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene 81, 245-257. |
| 33 | Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007). Clustal W and Clustal X version 2.0. Bio- informatics 23, 2947-2948. |
| 34 | Lynch M (2002). Genomics. Gene duplication and evolution. Science 297, 945-947. |
| 35 | Lynch M, Conery JS (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151-1155. |
| 36 | Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang H, Wang X, Bowers J, Paterson A, Lisch D, Freeling M (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol 148, 1772-1781. |
| 37 | Miché L, Belkin S, Rozen R, Balandreau J (2003). Rice seedling whole exudates and extracted alkylresorcinols induce stress-response in Escherichia coli biosensors. Environ Microbiol 5, 403-411. |
| 38 | Miyazono K, Um J, Imai FL, Katsuyama Y, Ohnishi Y, Horinouchi S, Tanokura M (2011). Crystal structure of curcuminoid synthase CUS from Oryza sativa. Proteins 79, 669-673. |
| 39 | Morita H, Wanibuchi K, Nii H, Kato R, Sugio S, Abe I (2010). Structural basis for the one-pot formation of the diarylheptanoid scaffold by curcuminoid synthase from Oryza sativa. Proc Natl Acad Sci USA 107, 19778-19783. |
| 40 | Parage C, Tavares R, Réty S, Baltenweck-Guyot R, Poutaraud A, Renault L, Heintz D, Lugan R, Marais GA, Aubourg S, Hugueney P (2012). Structural, func- tional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant Physiol 160, 1407-1419. |
| 41 | Proost S, Fostier J, De Witte D, Dhoedt B, Demeester P, Van de Peer Y, Vandepoele K (2012). i-ADHoRe 3.0-fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res 40, e11. |
| 42 | Rizzon C, Ponger L, Gaut BS (2006). Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol 2, e115. |
| 43 | Ross AB, Shepherd MJ, Schüpphaus M, Sinclair V, Alfaro B, Kamal-Eldin A, Aman P (2003). Alkylre- sorcinols in cereals and cereal products. J Agr Food Chem 51, 4111-4118. |
| 44 | Rubinstein CV, Gerrienne P, de la Puente GS, Astini RA, Steemans P (2010). Early Middle Ordovician evidence for land plants in Argentina (eastern Gondwana). New Phytol 188, 365-369. |
| 45 | Sanderson MJ (2003). Molecular data from 27 proteins do not support a Precambrian origin of land plants.Am J Bot 90, 954-956. |
| 46 | Schanz S, Schröder G, Schröder J (1992). Stilbene synthase from Scots pine (Pinus sylvestris). FEBS Lett 313, 71-74. |
| 47 | Schröder J (2000). The family of chalcone synthase-related proteins functional diversity and evolution.Recent Adv Phytochem 34, 55-89. |
| 48 | Schröder J (1997). A family of plant-specific polyketide synthases: facts and predictions. Trends Plant Sci 2, 373-378. |
| 49 | Schröder J, Schröder G (1990). Stilbene and chalcone synthases: related enzymes with key functions in plant- specific pathways. Z Naturforsch C 45, 1-8. |
| 50 | Stafford HA (1991). Flavonoid evolution: an enzymic ap- proach. Plant Physiol 96, 680-685. |
| 51 | Suyama M, Torrents D, Bork P (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34, W609-W612. |
| 52 | Tang H, Bowers JE, Wang X, Paterson AH (2010). Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci USA 107, 472-477. |
| 53 | Tropf S, Karcher B, Schröder G, Schröder J (1995). Reaction mechanisms of homodimeric plant polyketide synthases (stilbene and chalcone synthase). A single active site for the condensing reaction is sufficient for synthesis of stilbenes, chalcones, and 6'-deoxychalcon- es.J Biol Chem 270, 7922-7928. |
| 54 | Tropf S, Lanz T, Rensing SA, Schröder J, Schröder G (1994). Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. J Mol Evol 38, 610-618. |
| 55 | Vannozzi A, Dry IB, Fasoli M, Zenoni S, Lucchin M (2012). Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol 12, 130. |
| 56 | Vision TJ, Brown DG, Tanksley SD (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114-2117. |
| 57 | Walden AR, Walter C, Gardner RC (1999). Genes expressed in Pinus radiata male cones include homologs to anther-specific and pathogenesis response genes. Plant Physiol 121, 1103-1116. |
| 58 | Wang WK, Schaal BA, Chiou YM, Murakami N, Ge XJ, Huang CC, Chiang TY (2007). Diverse selective modes among orthologs/paralogs of the chalcone synthase (Chs) gene family of Arabidopsis thaliana and its relative A. halleri ssp. gemmifera. Mol Phylogenet Evol 44, 503-520. |
| 59 | Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009). Jalview Version 2―a multiple sequ- ence alignment editor and analysis workbench. Bioinfor- matics 25, 1189-1191. |
| 60 | Yamaguchi T, Kurosaki F, Suh DY, Sankawa U, Nishioka M, Akiyama T, Shibuya M, Ebizuka Y (1999). Cross- reaction of chalcone synthase and stilbene synthase overexpressed in Escherichia coli. FEBS Lett 460, 457-461. |
| 61 | Yang ZH (2007). PAML 4: phylogenetic analysis by maximum likelihood.Mol Biol Evol 24, 1586-1591. |
| 62 | Zarnowska ED, Zarnowski R, Kozubek A (2000). Alkyl- resorcinols in fruit pulp and leaves of Ginkgo biloba L. Z Naturforsch C 55, 881-885. |
| 63 | Zhan J (2009). Biosynthesis of bacterial aromatic poly- ketides. Curr Top Med Chem 9, 1958-1610. |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||