基因编辑技术在玉米中的研究进展
Research Progress of Gene Editing Technology in Maize
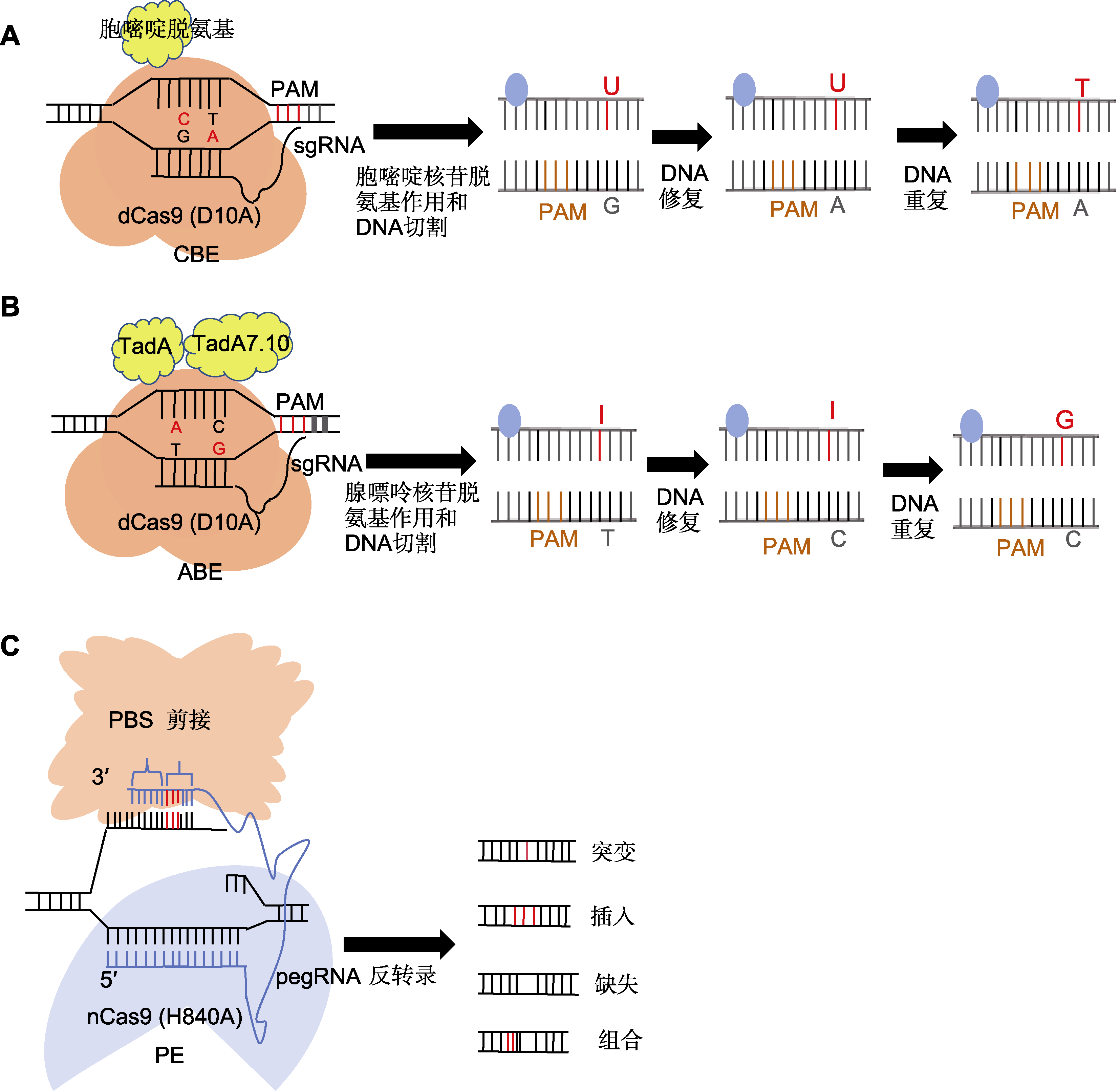
(A) sgRNA识别并结合到目标DNA序列特定的原间隔区邻近基序(PAM)位点, dCas9蛋白则与sgRNA结合, 形成复合物并结合于目标DNA上, 但不切割DNA双链, 仅使其单链化; 随后胞嘧啶脱氨酶在sgRNA的引导下, 接触到暴露的单链DNA上的胞嘧啶, 并催化其脱氨反应, 将胞嘧啶转化为尿嘧啶; 最后在DNA重复或修复过程中, 尿嘧啶被视为胸腺嘧啶的类似物, 从而纳入到新合成的DNA链中, 实现C-G碱基对到T-A碱基对的直接替换; (B) 利用CRISPR-Cas9系统中的sgRNA识别并结合到目标DNA序列的PAM位点, Cas9蛋白与sgRNA结合形成复合物, 并定位于目标DNA上; ABE中的腺嘌呤脱氨酶在sgRNA的引导下, 接触到暴露的单链DNA上的腺嘌呤, 并催化其脱氨反应, 将腺嘌呤转化为次黄嘌呤或脱氧次黄嘌呤; 最后在DNA修复过程中, 识别到次黄嘌呤后, 启动修复过程, 中间产物通常被替换为鸟嘌呤, 从而实现A-T碱基对到G-C碱基对的直接替换; (C) 利用pegRNA作为引导分子结合sgRNA, 并在其3'末端增加引物结合位点(PBS)序列和逆转录模板(RTT); 在pegRNA的引导下, 部分失活的Cas9切口酶切断含PAM序列的DNA单链; 切割后的DNA单链与pegRNA的3'末端PBS序列互补并结合, 随后逆转录酶沿RTT模板序列开始逆转录反应, 将目标编辑序列直接引到DNA切口处; 随后细胞内DNA修复机制识别并处理切口处的DNA结构, 最终保留携带目标编辑的DNA链。CBE、ABE和PE同
(A) The sgRNA recognizes and binds to the specific protospacer adjacent motif (PAM) site on the target DNA sequence, and the dCas9 protein binds to the sgRNA, forming a complex that attaches to the target DNA and rendering it single-stranded; subsequently, the cytidine deaminase, guided by the sgRNA, contacts the exposed cytidine on the single-stranded DNA and catalyzes its deamination, converting cytosine to uracil; finally, during DNA replication or repair, uracil is recognized as an analog of thymine and thymine is incorporated into the newly synthesized DNA strand, thereby achieving a direct replacement of the C-G base pair with a T-A base pair; (B) The sgRNA in the CRISPR-Cas9 system recognizes and binds to the PAM site on the target DNA sequence, and the Cas9 protein binds to the sgRNA to form a complex which is localized to the target DNA; under the direction of the sgRNA, adenine deaminase in the ABE system contacts the adenine on the single-stranded DNA, catalyzing its deamination reaction and converting adenine to hypoxanthine or deoxyhypoxanthine; during the DNA repair process, recognition of hypoxanthine triggers the initiation of the repair mechanism, typically resulting in the substitution of the intermediate product with guanine; this process facilitates the direct replacement of an A-T base pair with a G-C base pair; (C) Utilizing pegRNA as a guide molecule, it binds to the sgRNA and incorporates a primer binding site (PBS) sequence and reverse transcription template (RTT) at its 3' end; directed by the pegRNA, the partially deactivated Cas9 nickase cleaves the DNA single strand containing the PAM sequence; subsequently, the complementary PBS sequence at the 3' end of the pegRNA binds to the cleaved DNA strand; a reverse transcriptase then initiates a reverse transcription reaction along the RTT template sequence, directly incorporating the desired editing sequence into the DNA nick; finally, the intracellular DNA repair mechanisms are activated, ultimately retaining the DNA strand carrying the intended edit. CBE, ABE, and PE are the same as shown in