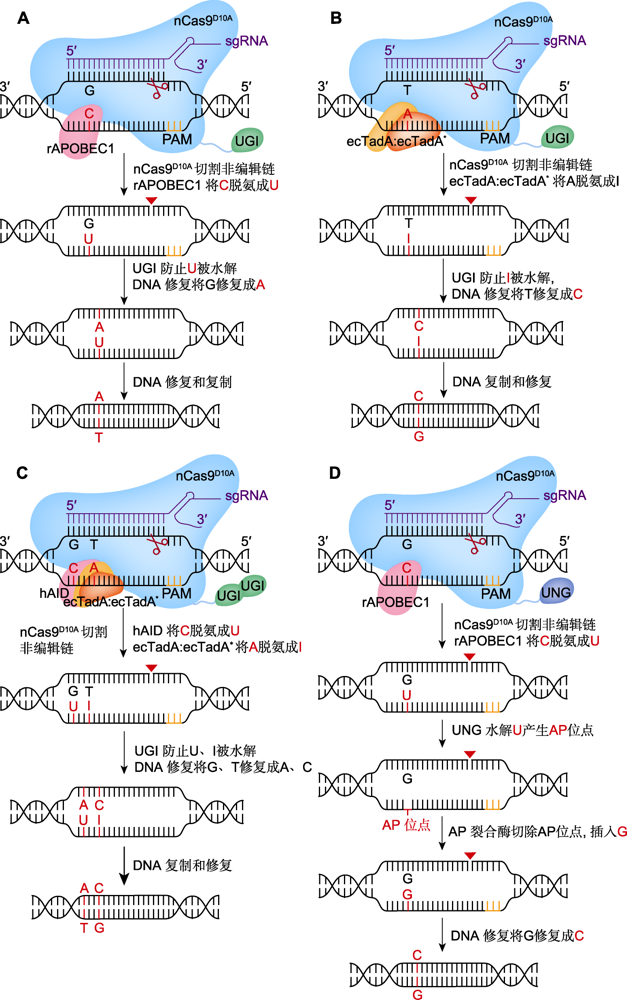
图1. 单(CBE、ABE、GBE)、双(A&C-BEmax)碱基编辑器工作原理
(A) BE3介导的C>T替换(在nCas9D10A (蓝色)、胞苷脱氨酶rAPOBEC1 (红色)和尿嘧啶糖基化酶抑制剂(UGI) (绿色)的作用下, 系统BE3将活性窗口中的C脱氨成U, 诱导细胞启动DNA修复实现C到T的替换; 红色三角形表示切口处); (B) ABE7.10介导的A>G替换(在腺苷脱氨酶ecTadA:ecTadA* (黄色和橙色)的作用下, 系统ABE7.10将活性窗口中的A脱氨成I, 并在DNA修复后实现A到G的替换); (C) A&C-BEmax介导的C>T与A>G共替换(在胞嘧啶脱氨酶hAID (红色)、ecTadA:ecTadA*和2个UGI的作用下, 系统A&C-BEmax诱导DNA修复, 实现C到T、A到G的同时替换); (D) GBE介导的C>G颠换(在尿嘧啶-N-糖基化酶(UNG) (深蓝)的作用下, UNG将C脱氨生成的U水解为AP位点, 并在DNA修复后实现C到G的颠换)。PAM: 原间隔序列邻近基序
Figure 1. Technical principles of single (CBE, ABE, GBE) and dual (A&C-BEmax) base editors
(A) BE3 mediated C to T base editing (mediated by nCas9D10A (blue), cytidine deaminase rAPOBEC1 (red) and uracil glycosylase inhibitor (UGI) (green), BE3 deaminates C in the active window into U and induces cells to start DNA repair and achieve the replacement of C to T; red triangle represents the notch); (B) ABE7.10 mediated A to G base editing (mediated by adenosine decease ecTadA:ecTadA* (yellow and orange), ABE7.10 deaminates A in the active window into I, and achieves A to G mutation after DNA repair); (C) A&C-BEmax mediated C to T and A to G base editing (mediated by cytidine deaminase hAID (red), ecTadA:ecTadA* and two UGI, A&C-BEmax induces DNA repair and achieves the simultaneous replacement of C to T and A to G); (D) GBE mediated C to G base editing (mediated by uracil-N-glycosylase (UNG) (deep blue), UNG hydrolyzes the U produced by C deamination to AP site, and achieves the transversion from C to G after DNA repair). PAM: Protospacer adjacent motif