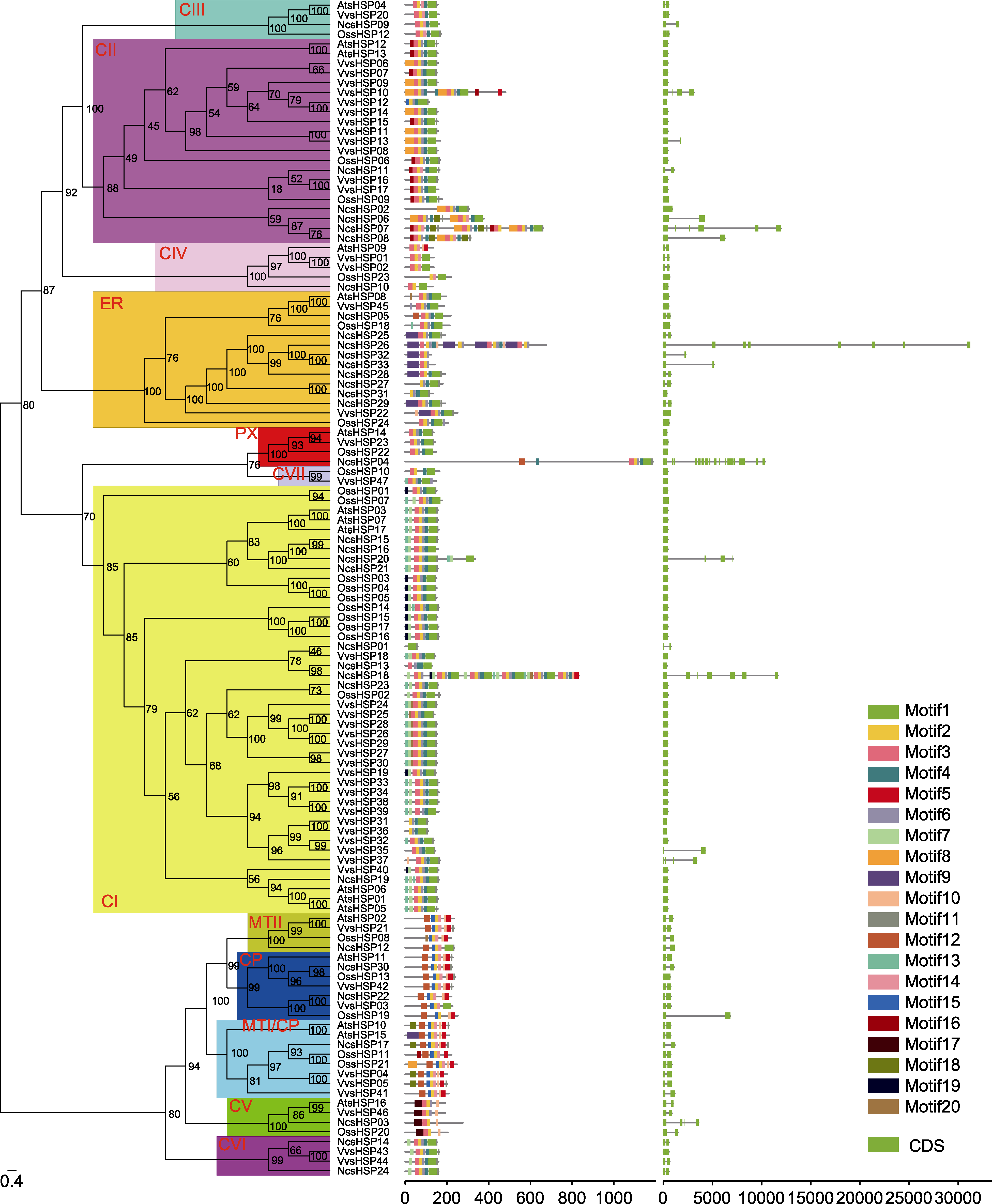
图2. 睡莲、水稻、拟南芥和葡萄中sHSP家族的系统进化(左)、保守基序(中)和基因结构(右)分析
通过IQ-tree软件构建系统进化树(左), 分支上的数字代表bootstrap值, sHSP亚家族用红色字母表示。保守基序(中)通过MEME程序识别, 不同颜色代表不同的保守基序, 保守基序的位置使用下方标尺进行估计。根据内含子和外显子的位置构建基因结构示意图(右), 绿色方框表示外显子, 黑色线表示内含子, 内含子和外显子的位置使用下方标尺进行估计。
Figure 2. The phylogenetic analysis (left), conserved motif (middle), and gene structure (right) of the sHSF family in waterlily, rice, Arabidopsis and grape
The phylogenetic tree (left) was constructed by IQ-tree software. The numbers in the clades stood for the bootstrap values, and sHSP subfamilies were indicated by red letters. The conserved motifs (middle) were identified by MEME software. Each conserved motif was marked by a specific color, and the location of motifs can be estimated using the scale at the bottom. The gene structures (right) were visualized according to the location of exons and introns. The exons and introns were represented by green box and black line, respectively. The location of exons and introns can be estimated using the scale at the bottom.