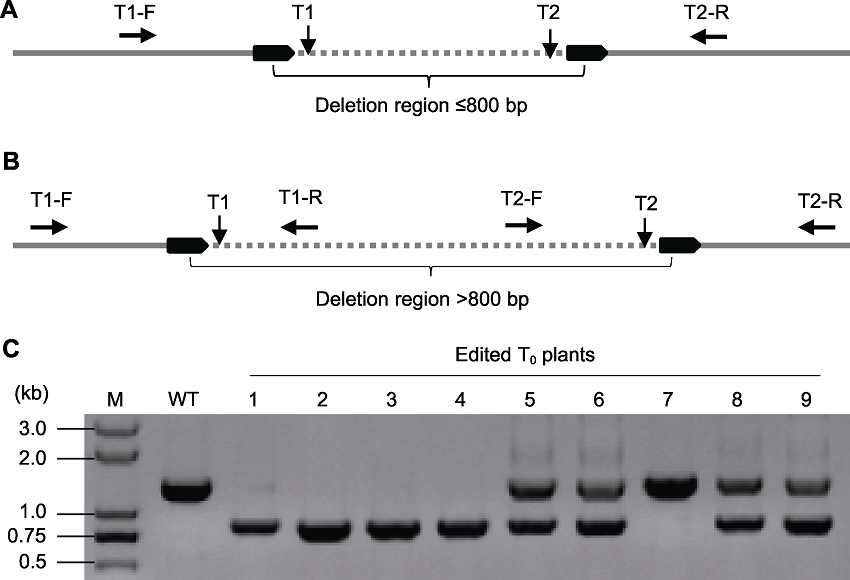
图3. 再生植株突变体突变类型的鉴定策略
(A) 如果删除片段小于或约等于800 bp, 在靶点1 (T1)的上游和靶点2 (T2)的下游设计正向引物(T1-F)和反向引物(T2-R)进行PCR扩增; (B) 如果删除片段大于800 bp, 合成4条引物(使以下各扩增产物长度相互有约70-100 bp的差别), 设3组PCR反应(T1-F/T1-R、T2-F/T2-R和T1-F/T2-R), 也可以在同一个PCR反应使用4条引物(如果只扩增出T1-F/T1-R片段和T2-F/T2-R片段而扩增不出T1-F/T2-R小片段, 表明没有发生片段删除; 如果只扩增出T1-F/T2-R小片段而没有扩增出T1-F/T1-R片段和T2-F/T2-R片段, 表明2个等位序列都产生了片段删除; 如果3个片段都被扩增出, 表明一个等位序列产生了片段删除而另一等位序列没有片段删除); (C) 按策略(A)的引物设计的PCR扩增电泳检测图。WT: 未编辑的野生型; 1-9为T0编辑植株, 其中植株1-4的2个等位序列都发生了片段删除; 植株5、6、8、9有1个等位序列发生了片段删除, 植株7未发生片段删除。
Figure 3. The strategy for identification of the mutant types in transgenic plants
(A) Amplification of the target region using the forward primer (T1-F) upstream the T1 and reverse primer (T2-R) downstream T2 if the length of desirable deleted fragment is less than 800 bp; (B) Four primers (with the length of the following amplification products is about 70-100 bp different from each other) with three combinations (T1-F/T1-R, T2-F/T2-R, and T1-F/T2-R) or pooled in an amplification are suggested if the length of desirable deleted fragment is more than 800 bp (there is no deletion of the target region if the resulting bands are produced with primer groups (T1-F/T1-R and T2-F/T2-R) but not T1-F/T2-R; both alleles had fragment deletion if the resulting bands are produced with primer group T1-F/T2-R but not T1-F/T1-R and T2-F/T2-R; one allele had fragment deletion and the other allele had no fragment deletion if all three fragments were produced; (C) The example of gel electrophoresis according to the detection method of (A). WT: Wild type; 1-9 indicate edited T0 plants in which 1-4 with biallelic fragment deletion, 5, 6, 8 and 9 with heterozygous deletion and 7 with no deletion.