Recent Uplift of the Taihang Mountains Triggered the Lineage Diversification within the Genus Taihangia (Rosaceae)
- 1College of Landscape and Ecological Engineering, Hebei University of Engineering, Handan 056038, China
2College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou 510642, China
Received date: 2023-11-29
Accepted date: 2024-05-04
Online published: 2024-05-07
Abstract
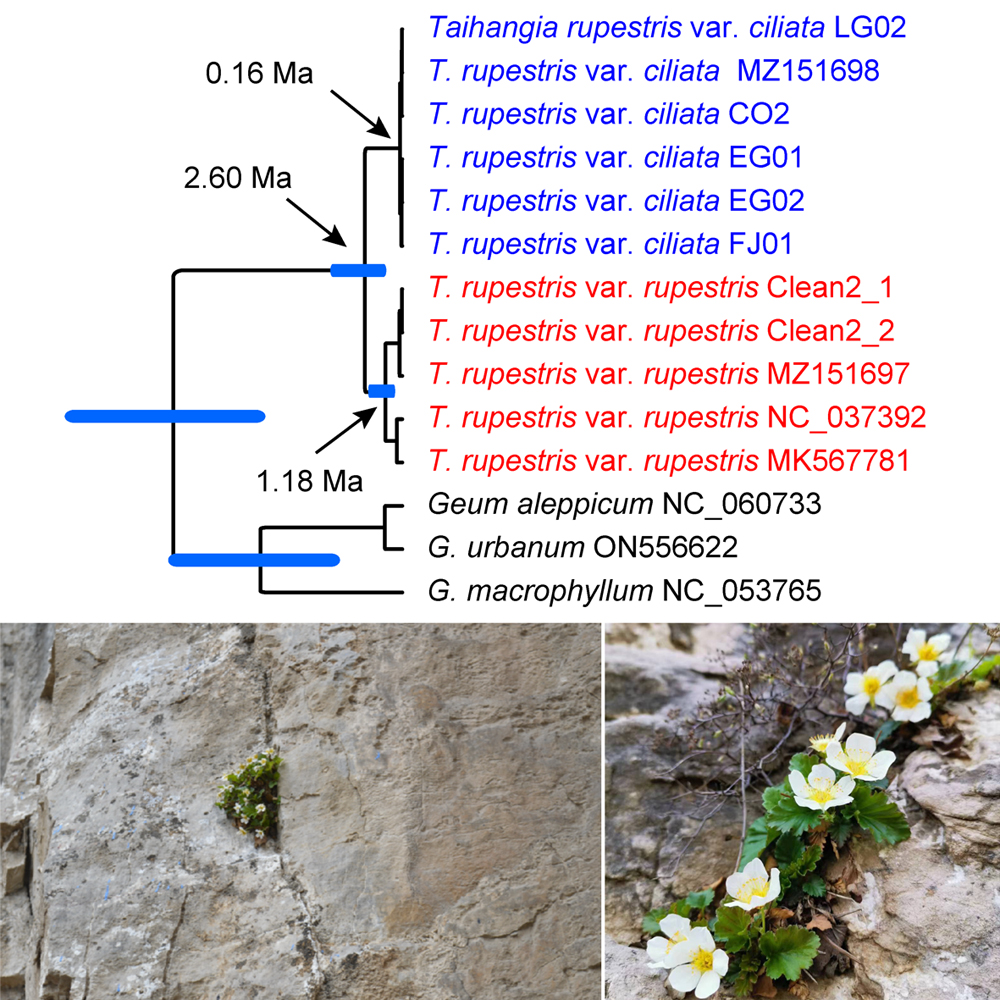
Taihangia is a monotypic genus of the Rosaceae and endemic to the southern part of Taihang Mountains. Two varieties (T. rupestris var. rupestris and T. rupestris var. ciliate) are circumscribed currently under the species T. rupestris. However, the taxonomic status of these two varieties is still controversial and very few studies on the evolutionary history of this genus. In this study, a plastid phylogenomic analysis of Taihangia was conducted and the temporal evolutionary history of the genus was investigated. The results showed that the monophyly of the genus and also the two varieties were all recovered with strong support. In addition, the genus started to diverge at ca. 2.60 million years ago (Ma) near the Pliocene-Pleistocene boundary, and diversification events within the two varieties were estimated mostly during the late Pleistocene, which is highly consistent in time scale with the recent uplift of the southern part of the Taihang Mountains that occurred during the Pliocene and Pleistocene. Thus, we propose that the uplift of the southern part of Taihang Mountains may have played an important role in triggering the lineage diversification within the genus Taihangia. The present study not only enhances our understanding on the evolutionary history of Taihangia, but also provides a case study in understanding the relationship between diversification of plant lineages and mountains uplifting occurred in Asia.
Key words: divergence times; Rosaceae; mountain uplifting; Taihangia; Taihang Mountains; phylogenomics
Cite this article
Wenna Chen , Liangtao Li , Lu Zhou , Gang Yao . Recent Uplift of the Taihang Mountains Triggered the Lineage Diversification within the Genus Taihangia (Rosaceae)[J]. Chinese Bulletin of Botany, 2024 , 59(5) : 763 -773 . DOI: 10.11983/CBB23159
References
| [1] | Bolger AM, Lohse M, Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. |
| [2] | Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10, e1003537. |
| [3] | Chen X, Li JL, Cheng T, Zhang W, Liu YL, Wu P, Yang XY, Wang L, Zhou SL (2020). Molecular systematics of Rosoideae (Rosaceae). Plant Syst Evol 306, 9. |
| [4] | Cheng YQ, Duan JM, Jiao ZB, Wang GG, Yan FM, Wang HW (2016). Cytoplasmic DNA disclose high nucleotide diversity and different phylogenetic pattern in Taihangia rupestris Yu et Li. Biochem Syst Ecol 66, 201-208. |
| [5] | Ding WN, Ree RH, Spicer RA, Xing YW (2020). Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science 369, 578-581. |
| [6] | Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19, 11-15. |
| [7] | Duan JM (2013). The Research on Genetic Diversify and Molecular Phylogeography Population of Taihangia rupestris Yu et Li. Master’s thesis. Zhengzhou: Henan Agricultural University. pp. 1-45. (in Chinese) |
| 段景勉 (2013). 太行花种群遗传多样性与分子谱系地理学研究. 硕士论文. 郑州: 河南农业大学. pp. 1-45. | |
| [8] | Edelman DW (1975). The Eocene Germer Basin Flora of South-central Idaho. Master’s thesis. Moscow: University of Idaho. pp. 1-142. |
| [9] | Fang XM (2010). Genetic Diversity Study of Rare and Precious Species Taihangia rupestris. Master’s thesis. Zhengzhou: Henan Agricultural University. pp. 1-48. (in Chinese) |
| 方向民 (2010). 珍稀植物太行花的遗传多样性研究. 硕士论文. 郑州: 河南农业大学. pp. 1-48. | |
| [10] | Favre A, Michalak I, Chen CH, Wang JC, Pringle JS, Matuszak S, Sun H, Yuan YM, Struwe L, Muellner- Riehl AN (2016). Out-of-Tibet: the spatio-temporal evolution of Gentiana (Gentianaceae). J Biogeogr 43, 1967-1978. |
| [11] | Feng Z, Zheng Y, Jiang Y, Li LZ, Luo GM, Huang LF (2022). The chloroplast genomes comparative analysis of Taihangia rupestris var. rupestris and Taihangia rupestris var. ciliate, two endangered and endemic cliff plants in Tai- hang Mountain of China. South Afr J Bot 148, 499-509. |
| [12] | Geng X, Ye J, Jian YH, Zhang HM (2009). Advances on the study of special rare and endangered plant Taihangia rupestris in China. J Anhui Agri Sci 35, 8965-8966, 8973. (in Chinese) |
| 耿霄, 叶嘉, 焦云红, 张会敏 (2009). 中国特有珍稀濒危植物太行花的研究进展. 安徽农业科学 35, 8965-8966, 8973. | |
| [13] | He WC, Chen CJ, Xiang KL, Wang J, Zheng P, Tembrock LR, Jin DM, Wu ZQ (2021). The history and diversity of rice domestication as resolved from 1464 complete plastid genomes. Front Plant Sci 12, 781793. |
| [14] | Hou ZG, Li JB, Li SQ (2014). Diversi?cation of low dispersal crustaceans through mountain uplift: a case study of Gammarus (Amphipoda: Gammaridae) with descriptions of four novel species. Zool J Linn Soc 170, 591-633. |
| [15] | Hughes CE, Atchison GW (2015). The ubiquity of alpine plant radiations: from the Andes to the Hengduan Mountains. New Phytol 207, 275-282. |
| [16] | Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ (2020a). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol 21, 241. |
| [17] | Jin WY, Li HW, Wei R, Huang BH, Liu B, Sun TT, Mabberley DJ, Liao PC, Yang Y (2020b). New insights into biogeographical disjunctions between Taiwan and the Eastern Himalayas: the case of Prinsepia (Rosaceae). Ta- xon 69, 278-289. |
| [18] | Katoh K, Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772-780. |
| [19] | Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647-1649. |
| [20] | Li WG, Zhang LH, Zhang YD, Wang GD, Song DY, Zhang YW (2017). Selection and validation of appropriate reference genes for quantitative real-time PCR normalization in staminate and perfect flowers of andromonoecious Taihangia rupestris. Front Plant Sci 8, 729. |
| [21] | Lin N, Landis JB, Sun YX, Huang XH, Zhang X, Liu Q, Zhang HJ, Sun H, Wang HC, Deng T (2021). Demographic history and local adaptation of Myripnoisdioica (Asteraceae) provide insight on plant evolution in northern China flora. Ecol Evol 11, 8000-8013. |
| [22] | Ma YS, Zhao X, Zhao XT, Wu ZH, Gao LZ, Zhang YQ, Zhao T, Wu ZH, Yang SZ (2007). The Cenozoic rifting and uplifting process on the Southern margin of Taihangshan uplift. Acta Geoscient Sin 28, 219-233. (in Chinese) |
| 马寅生, 赵逊, 赵希涛, 吴中海, 高林志, 张岳桥, 赵汀, 吴珍汉, 扬守政 (2007). 太行山南缘新生代的隆升与断陷过程. 地球学报 28, 219-233. | |
| [23] | Posada D (2008). jModelTest: phylogenetic model averaging. Mol Biol Evol 25, 1253-1256. |
| [24] | Protopopova M, Pavlichenko V, Chepinoga V, Gnutikov A, Adelshin R (2023). Waldsteinia within Geums. l. (Rosaceae): main aspects of phylogeny and speciation history. Diversity 15, 479. |
| [25] | Qin M, Zhu CJ, Yang JB, Vatanparast M, Schley R, Lai Q, Zhang DY, Tu TY, Klitgård BB, Li SJ, Zhang DX (2022). Comparative analysis of complete plastid genome reveals powerful barcode regions for identifying wood of Dalbergia odorifera and D. tonkinensis (Leguminosae). J Syst Evol 60, 73-84. |
| [26] | Qu XJ, Moore MJ, Li DZ, Yi TS (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 50. |
| [27] | Rambaut A, Suchard MA, Drummond AJ (2014). Tracer version 1. 6. http://beast.bio.ed.ac.uk/Tracer. 2022-08-10. |
| [28] | Ronquist F, Huelsenbeck JP (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574. |
| [29] | Smedmark JEE (2006). Recircumscription of Geum (Colurieae: Rosaceae). Bot Jahrb Syst 126, 409-417. |
| [30] | Smedmark JEE, Eriksson T (2002). Phylogenetic relationships of Geum (Rosaceae) and relatives inferred from the nrITS and trnL-trnF regions. Syst Bot 27, 303-317. |
| [31] | Stamatakis A (2006). RAxML-VI-HPC: maximum likelihood- based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688-2690. |
| [32] | Sun X, Wang YP, Liu C, Huang LF (2019). Molecular identification of Taihangia rupestris Yu et Li, an endangered species endemic to China. South Afr J Bot 124, 173-177. |
| [33] | Tang M (2004). Conservation Biology of a Rare Herb Taihangia rupestris ( Rosaceae). PhD dissertation. Beijing: Graduate School of the Chinese Academy of Sciences (Institute of Botany). pp.1-127. (in Chinese) |
| 唐敏 (2004). 稀有植物太行花的保护生物学研究. 博士论文. 北京: 中国科学院研究生院(植物研究所). pp. 1-127. | |
| [34] | Tang M, Yu FH, Jin XB, Ge S (2010). High genetic diversity in the naturally rare plant Taihangia rupestris Yu et Li (Rosaceae) dwelling only cliff faces. Pol J Ecol 58, 241- 248. |
| [35] | Tang WB (2005). A floristic analysis of the seed plants in the middle-south area of east slope of Taihang Mountains. Bull Bot Res 25, 366-372. (in Chinese) |
| 唐伟斌 (2005). 太行山脉东坡中南段种子植物区系初步分析. 植物研究 25, 366-372. | |
| [36] | Vozárová R, Herklotz V, Kovařík A, Tynkevich YO, Volkov RA, Ritz CM, Lunerová J (2021). Ancient origin of two 5S rDNA families dominating in the genus Rosa and their behavior in the canina-type meiosis. Front Plant Sci 12, 643548. |
| [37] | Wang HW, Cheng YQ, Fang XM, Ye YZ, Zong YY, Cao RZ (2011). Genetic diversity and differentiation in rare herb species Taihangia rupestris (Rosaceae). Acta Bot Boreal-Occident Sin 31, 45-51. (in Chinese) |
| 王红卫, 程月琴, 方向民, 叶永忠, 宗盈盈, 曹荣哲 (2011). 珍稀植物太行花的遗传多态性及其分化. 西北植物学报 31, 45-51. | |
| [38] | Wang HW, Fang XM, Ye YZ, Cheng YQ, Wang ZS (2011). High genetic diversity in Taihangia rupestris Yu et Li, a rare cliff herb endemic to China, based on inter-simple sequence repeat markers. Biochem Syst Ecol 39, 553-561. |
| [39] | Wang JS (2008). Research review Taihangia rupestris as a wild ornamental. J Anhui Agric Sci 36, 6209-6210. (in Chinese) |
| 王建书 (2008). 野生珍稀观赏植物太行花的研究. 安徽农业科学 36, 6209-6210. | |
| [40] | Wang YL, Yan GQ (2014). Molecular phylogeography and population genetic structure of O. longilobus and O. taihangensis (Opisthopappus) on the Taihang Mountains. PLoS One 9, e104773. |
| [41] | Wu C, Zhang XQ, Ma YH (1999). The Taihang and Yan Mountains rose mainly in Quarteranary. North China Earthq Sci 17, 1-7. (in Chinese) |
| 吴忱, 张秀清, 马永红 (1999). 太行山、燕山主要隆起于第四纪. 华北地震科学 17, 1-7. | |
| [42] | Xu GF, Li XW, Meng L, Wang HS (2006). Resources of main varieties of wild herbal flower of Taihang mountain regions in Henan. J Anhui Agri Sci 34, 2380-2381, 2507. (in Chinese) |
| 许桂芳, 李孝伟, 孟丽, 王鸿升 (2006). 河南太行山区主要野生草本花卉资源. 安徽农业科学 34, 2380-2381, 2507. | |
| [43] | Xue C, Geng FD, Li JJ, Zhang DQ, Gao F, Huang L, Zhang XH, Kang JQ, Zhang JQ, Ren Y (2021). Divergence in the Aquilegia ecalcarata complex is correlated with geography and climate oscillations: evidence from plastid genome data. Mol Ecol 30, 5796-5813. |
| [44] | Yao G, Zhang YQ, Barrett C, Xue BE, Bellot S, Baker WJ, Ge XJ (2023). A plastid phylogenomic framework for the palm family (Arecaceae). BMC Biol 21, 50. |
| [45] | Ye H, Wang Z, Hou HM, Wu JH, Gao Y, Han W, Ru WM, Sun GL, Wang YL (2021). Localized environmental heterogeneity drives the population differentiation of two endangered and endemic Opisthopappus Shih species. BMC Ecol Evol 21, 56. |
| [46] | Ye XY, Ma PF, Yang GQ, Guo C, Zhang YX, Chen YM, Guo ZH, Li DZ (2019). Rapid diversification of alpine bamboos associated with the uplift of the Hengduan Moun- tains. J Biogeogr 46, 2678-2689. |
| [47] | Yü TT, Li CL (1980). Taihangia Yu et Li—a new genus of Rosaceae from China. J Univ Chin Acad Sci 18, 469-472. (in Chinese) |
| 俞德浚, 李朝銮 (1980). 太行花属——蔷薇科一新属. 中国科学院大学学报 18, 469-472. | |
| [48] | Yü TT, Li CL (1983). The systematic position of genus Taihangia in Rosaceae. J Univ Chin Acad Sci 21, 229-235. (in Chinese) |
| 俞德浚, 李朝銮 (1983). 蔷薇科太行花属系统位置的研究. 中国科学院大学学报 21, 229-235. | |
| [49] | Yu XQ, Maki M, Drew BT, Paton AJ, Li HW, Zhao JL, Conran JG, Li J (2014). Phylogeny and historical biogeography of Isodon (Lamiaceae): rapid radiation in south- west China and Miocene overland dispersal into Africa. Mol Phylogenet Evol 77, 183-194. |
| [50] | Zhang JQ, Meng SY, Allen GA, Wen J, Rao GY (2014). Rapid radiation and dispersalout of the Qinghai-Tibetan plateau of an alpine plant lineage Rhodiola (Crassulaceae). Mol Phylogenet Evol 77, 147-158. |
| [51] | Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS (2017). Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol 214, 1355-1367. |
| [52] | Zhao F, Chen YP, Salmaki Y, Drew BT, Wilson TC, Scheen AC, Celep F, Bräuchler C, Bendiksby M, Wang Q, Min DZ, Peng H, Olmstead RG, Li B, Xiang CL (2021). An updated tribal classification of lamiaceae based on plastome phylogenomics. BMC Biol 19, 2. |
| [53] | Zhao Q, Liu HX, Luo LG, Ji X (2011). Comparative population genetics and phylogeography of two lacertid lizards (Eremias argus and E. brenchleyi) from China. Mol Phylogenet Evol 58, 478-491. |
| [54] | Zhao X, Gao CW (2020). The complete chloroplast genome sequence of Rosa minutifolia. Mitochondrial DNA Part B 5, 3320-3321. |
| [55] | Zhou Z, Hong DY, Niu Y, Li GD, Nie ZL, Wen J, Sun H (2013). Phylogenetic and biogeographic analyses of the sino-Himalayan endemic genus Cyananthus (Campanulaceae) and implications for the evolution of its sexual system. Mol Phylogenet Evol 68, 482-497. |