Regulation of Plasma Membrane Protein Dynamics and Its Research Methods
- 1. State Key Laboratory of Tree Genetics and Breeding, College of Biological Sciences and Technology, Beijing Forestry University, Beijing 100083, China
2. National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, Beijing 100083, China
3. Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, College of Biological Sciences and Technology, Beijing Forestry University, Beijing 100083, China
4. The Tree and Ornamental Plant Breeding and Bioengineering Key Laboratory of National Forestry and Grassland Administration, Beijing Forestry University, Beijing 100083, China
5. Natural Resources and Planning Bureau of Weishan County, Jining 277600, China
Received date: 2022-06-06
Accepted date: 2022-11-12
Online published: 2022-11-29
Abstract
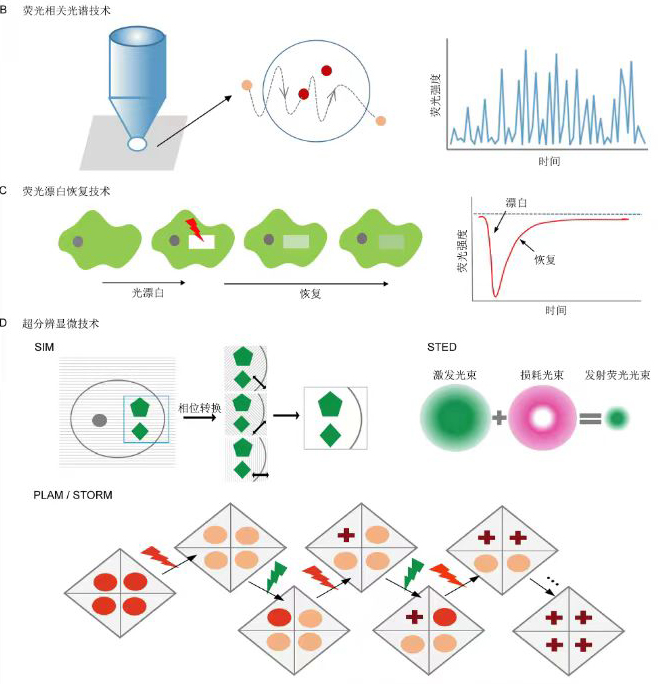
Plasma membrane (PM) proteins are important components of cell membranes and play important roles in material transport, ion exchange, signal transduction, and metabolic process. Their movements in the PM are in response to the developmental cues and environmental stimuli. Studying the regulatory mechanism of PM protein movement is crucial for a better understanding of the plant development and adaptation to environment. In the recent years, the rapid development of microscopic technologies enables us to move one step closer to reveal the regulatory mechanism of PM protein dynamics. In this paper, we systematically summarized PM protein dynamics and the factors affecting it. We also provided an introduction to commonly-used microscopic imaging techniques applied on PM protein dynamics research. This review will serve as a useful reference for the further investigation of biological functions of PM proteins.
Key words: plasma membrane protein; dynamics; microdomain; single-molecule imaging
Cite this article
Pengyun Luo , Hongping Qian , Yan Liu , Changwen Xu , Yaning Cui . Regulation of Plasma Membrane Protein Dynamics and Its Research Methods[J]. Chinese Bulletin of Botany, 2023 , 58(4) : 590 -601 . DOI: 10.11983/CBB22102
References
| [1] | 李晓娟 (2012). 拟南芥质膜水通道蛋白活性调控多重机制的单分子研究. 博士论文. 北京: 中国科学院大学. pp. 1-93. |
| [2] | 刘玥, 尹悦佳, 梁重阳, 黄殿帅, 王阳, 刘艳芝, 窦瑶, 冯树丹, 郝东云 (2015). 3D-SIM结构照明超分辨率显微镜实现蛋白质在植物亚细胞器内的定位. 植物学报 50, 495-503. |
| [3] | 钱虹萍, 陈博, 林金星, 崔亚宁 (2021). RNA聚合酶II动态调控及其成像技术的研究进展. 生物技术通报 37, 293-302. |
| [4] | 邱丽丽, 赵琪, 张玉红, 戴绍军 (2017). 植物质膜蛋白质组的逆境应答研究进展. 植物学报 52, 128-147. |
| [5] | 苏泊丹, 林金星, 肖建伟 (2016). 泛素介导的植物膜蛋白转运及其研究方法. 植物学报 51, 387-395. |
| [6] | Ananthanarayanan V (2021). Cytoskeletal mechanics. J Indian Inst Sci 101, 3. |
| [7] | Chu TTH, Hoang TG, Trinh DC, Bureau C, Meynard D, Vernet A, Ingouff M, Do NV, Périn C, Guiderdoni E, Gantet P, Maurel C, Luu DT (2018). Sub-cellular markers highlight intracellular dynamics of membrane proteins in response to abiotic treatments in rice. Rice (NY) 11, 23. |
| [8] | Cui YN, Li XJ, Yu M, Li RL, Fan LS, Zhu YF, Lin JX (2018a). Sterols regulate endocytic pathways during flg22- induced defense responses in Arabidopsis. Development 145, dev165688. |
| [9] | Cui YN, Yu M, Yao XM, Xing JJ, Lin JX, Li XJ (2018b). Single-particle tracking for the quantification of membrane protein dynamics in living plant cells. Mol Plant 11, 1315-1327. |
| [10] | Cui YN, Zhao YX, Lu YQ, Su X, Chen YY, Shen YB, Lin JX, Li XJ (2021). In vivo single-particle tracking of the aquaporin AtPIP2;1 in stomata reveals cell type-specific dynamics. Plant Physiol 185, 1666-1681. |
| [11] | Donnert G, Keller J, Medda R, Andrei MA, Rizzoli SO, Lührmann R, Jahn R, Eggeling C, Hell SW (2006). Macromolecular-scale resolution in biological fluorescence microscopy. Proc Natl Acad Sci USA 103, 11440-11445. |
| [12] | Douglass AD, Vale RD (2005). Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell 121, 937-950. |
| [13] | Gutierrez R, Grossmann G, Frommer WB, Ehrhardt DW (2010). Opportunities to explore plant membrane organization with super-resolution microscopy. Plant Physiol 154, 463-466. |
| [14] | Hao HQ, Fan LS, Chen T, Li RL, Li XJ, He QH, Botella MA, Lin JX (2014). Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell 26, 1729-1745. |
| [15] | Hoffmann N, King S, Samuels AL, McFarlane HE (2021). Subcellular coordination of plant cell wall synthesis. Dev Cell 56, 933-948. |
| [16] | Jaqaman K, Grinstein S (2012). Regulation from within: the cytoskeleton in transmembrane signaling. Trends Cell Biol 22, 515-526. |
| [17] | Jozefowicz AM, Matros A, Witzel K, Mock HP (2018). Mini-scale isolation and preparation of plasma membrane proteins from potato roots for LC/MS analysis. In: Mock HP, Matros A, Witzel K, eds. Plant Membrane Proteomics. New York: Humana Press. pp. 195-204. |
| [18] | Kipper FC, Tamajusuku ASK, Minussi DC, Vargas JE, Battastini AMO, Kaczmarek E, Robson SC, Lenz G, Wink MR (2018). Analysis of NTPDase2 in the cell membrane using fluorescence recovery after photobleaching (FRAP). Cytometry A 93, 232-238. |
| [19] | Kleine-Vehn J, Wabnik K, Martinière A, ?angowski ?, Willig K, Naramoto S, Leitner J, Tanaka H, Jakobs S, Robert S, Luschnig C, Govaerts W, Hell SW, Runions J, Friml J (2011). Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol 7, 540. |
| [20] | Komis G, Mistrik M, ?amajová O, Dosko?ilová A, Ove?ka M, Illés P, Bartek J, ?amaj J (2014). Dynamics and organization of cortical microtubules as revealed by superresolution structured illumination microscopy. Plant Physiol 165, 129-148. |
| [21] | Kure JL, Andersen CB, Mortensen KI, Wiseman PW, Arnspang EC (2020). Revealing plasma membrane nano-domains with diffusion analysis methods. Membranes (Basel) 10, 314. |
| [22] | Le Gall H, Philippe F, Domon JM, Gillet F, Pelloux J, Rayon C (2015). Cell wall metabolism in response to abiotic stress. Plants (Basel) 4, 112-166. |
| [23] | Li XJ, Luu DT, Maurel C, Lin JX (2013). Probing plasma membrane dynamics at the single-molecule level. Trends Plant Sci 18, 617-624. |
| [24] | Li XJ, Wang XH, Yang Y, Li RL, He QH, Fang XH, Luu DT, Maurel C, Lin JX (2011). Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23, 3780-3797. |
| [25] | Lv XQ, Jing YP, Xiao JW, Zhang YD, Zhu YF, Julian R, Lin JX (2017). Membrane microdomains and the cytoskeleton constrain AtHIR1 dynamics and facilitate the formation of an AtHIR1-associated immune complex. Plant J 90, 3-16. |
| [26] | Machta BB, Papanikolaou S, Sethna J, Veatch S (2011). Minimal model of plasma membrane heterogeneity requires coupling cortical actin to criticality. Biophys J 100, 1668-1677. |
| [27] | Malinovsky FG, Fange lJU, Willats WGT (2014). The role of the cell wall in plant immunity. Front Plant Sci 5, 178. |
| [28] | Martínez-Mu?oz L, Rodriguez-Frade JM, Barroso R, Sorzano CóS, Torreno-Pina JA, Sa?tiago CA, Manzo C, Lucas P, García-Cuesta EM, Gutierrez E, Barrio L, Vargas J, Cascio G, Carrasco YR, Sánchez-Madrid F, García-Parajo MF, Mellado M (2018). Separating actin-dependent chemokine receptor nanoclustering from dimerization indicates a role for clustering in CXCR4 signaling and function. Mol Cell 70, 106-119. |
| [29] | Martinière A, Lavagi I, Nageswaran G, Rolfe DJ, Maneta- Peyret L, Luu DT, Botchway SW, Webb SED, Mongrand S, Maurel C, Martin-Fernandez ML, Kleine-Vehn J, Friml J, Moreau P, Runions J (2012). Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc Natl Acad Sci USA 109, 12805-12810. |
| [30] | Mattila PK, Batista FD, Treanor B (2016). Dynamics of the actin cytoskeleton mediates receptor cross talk: an emerging concept in tuning receptor signaling. J Cell Biol 212, 267-280. |
| [31] | McKenna JF, Rolfe DJ, Webb SED, Tolmie AF, Botchway SW, Martin-Fernandez ML, Hawes C, Runions J (2019). The cell wall regulates dynamics and size of plasma-membrane nanodomains in Arabidopsis. Proc Natl Acad Sci USA 116, 12857-12862. |
| [32] | Mudumbi KC, Schirmer EC, Yang WD (2016). Single-point single-molecule FRAP distinguishes inner and outer nuclear membrane protein distribution. Nat Commun 7, 12562. |
| [33] | Mudumbi KC, Yang WD (2017). Determination of membrane protein distribution on the nuclear envelope by single-point single-molecule FRAP. Curr Protoc Cell Biol 76, 21.11.1-21.11.13. |
| [34] | Nederveen-Schippers LM, Pathak P, Keizer-Gunnink I, Westpha lAH, Van Haastert PJM, Borst JW, Kortholt A, Skakun V (2021). Combined FCS and PCH analysis to quantify protein dimerization in living cells. Int J Mol Sci 22, 7300. |
| [35] | Pinaud F, Michalet X, Iyer G, Margeat E, Moore HP, Weiss S (2009). Dynamic partitioning of a glycosyl-phosphatidylinositol-anchored protein in glycosphingolipid-rich microdomains imaged by single-quantum dot tracking. Traffic 10, 691-712. |
| [36] | Rostoks N, Schmierer D, Kudrna D, Kleinhofs A (2003). Barley putative hypersensitive induced reaction genes: genetic mapping, sequence analyses and differential expression in disease lesion mimic mutants. Theor Appl Genet 107, 1094-1101. |
| [37] | Schneider F, Hernandez-Varas P, Lagerholm BC, Shrestha D, Sezgin E, Roberti MJ, Ossato G, Hecht F, Eggeling C, Urban?i? I (2020). High photon count rates improve the quality of super-resolution fluorescence fluctuation spectroscopy. J Phys D Appl Phys 53, 164003. |
| [38] | Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, Vorwerk S, Youngs H (2004). Toward a systems approach to understanding plant cell walls. Science 306, 2206-2211. |
| [39] | Son S, Oh CJ, An CS (2014). Arabidopsis thaliana remorins interact with SnRK1 and play a role in susceptibility to beet curly top virus and beet severe curly top virus. Plant Pathol J 30, 269-278. |
| [40] | Su BD, Zhang X, Li L, Abbas S, Yu M, Cui YN, Balu?ka F, Hwang I, Shan XY, Lin JX (2021). Dynamic spatial reorganization of BSK1 complexes in the plasma membrane underpins signal-specific activation for growth and immunity. Mol Plant 14, 588-603. |
| [41] | Sundd P, Gutierrez E, Pospieszalska M, Groisman A, Ley K (2010). Stressed and compressed molecular bonds revealed in footprints of rolling neutrophils using total internal reflection fluorescence microscopy. Biophys J 98, 595a. |
| [42] | Tingey M, Li YC, Yang WD (2021). Protocol for single-molecule fluorescence recovery after photobleaching microscopy to analyze the dynamics and spatial locations of nuclear transmembrane proteins in live cells. STAR Pro-toc 2, 100490. |
| [43] | Villarruel C, Dawson SP (2020). Quantification of fluctua-tions from fluorescence correlation spectroscopy experi-ments in reaction-diffusion systems. Phys Rev E 102, 052407. |
| [44] | Wan YL, Ash III WM, Fan LS, Hao HQ, Kim MK, Lin JX (2011). Variable-angle total internal reflection fluorescence microscopy of intact cells of Arabidopsis thaliana. Plant Methods 7, 27. |
| [45] | Wang L, Li H, Lv XQ, Chen T, Li RL, Xue YQ, Jiang JJ, Jin B, Balu?ka F, ?amaj J, Wang XL, Lin JX (2015). Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living Arabidopsis cells. Mol Plant 8, 1334-1349. |
| [46] | Wang QL, Zhao YY, Luo WX, Li RL, He QH, Fang XH, Michele RD, Ast C, Von Wirén N, Lin JX (2013). Single- particle analysis reveals shutoff control of the Arabidopsis ammonium transporter AMT1;3 by clustering and inter-nalization . Proc Natl Acad Sci USA 110, 13204-13209. |
| [47] | Wohland T, Bag N, Ma XX, Huang SR (2013). Epidermal growth factor receptor localization in cell membranes in-vestigated by imaging FCS. Biophys J 104, 681a. |
| [48] | Wollert T, Langford GM (2022). Super-resolution imaging of the actin cytoskeleton in living cells using TIRF-SIM. In: Gavin RH, ed. Cytoskeleton. New York: Humana. pp. 3-24. |
| [49] | Xing JJ, Li XJ, Wang XH, Lv XQ, Wang L, Zhang L, Zhu YF, Shen QH, Balu?ka F, ?amaj J, Lin JX (2019). Se-cretion of phospholipase D deltafunctions as a regulatory mechanism in plant innate immunity. Plant Cell 31, 3015-3032. |
| [50] | Xu CW, Abbas S, Qian HP, Yu M, Zhang X, Li XJ, Cui YN, Lin JX (2022). Environmental cues contribute to dynamic plasma membrane organization of nanodomains containing flotillin-1 and hypersensitive induced reaction-1 pro-teins in Arabidopsis thaliana. Front Plant Sci 13, 897594. |
| [51] | Xue YQ, Xing JJ, Wan YL, Lv XQ, Fan LS, Zhang YD, Song K, Wang L, Wang XH, Deng X, Balu?ka F, Christie JM, Lin JX (2018). Arabidopsis blue light receptor phototropin 1 undergoes blue light-induced activation in membrane microdomains. Mol Plant 11, 846-859. |
| [52] | Yasui M, Hiroshima M, Kozuka J, Sako Y, Ueda M (2018). Automated single-molecule imaging in living cells. Nat Commun 9, 3061. |
| [53] | Yu M, Li RL, Cui YN, Chen WJ, Li B, Zhang X, Bu YF, Cao YY, Xing JJ, Jewaria PK, Li XJ, Bhalerao RP, Yu F, Lin JX (2020). The RALF1-FERONIA interaction modulates endocytosis to mediate control of root growth in Arabidop-sis. Development 147, dev189902. |
| [54] | Yu M, Liu HJ, Dong ZY, Xiao JW, Su BD, Fan LS, Komis G, ?amaj J, Lin JX, Li RL (2017). The dynamics and endocytosis of Flot1 protein in response to flg22 in Arabidopsis. J Plant Physiol 215, 73-84. |
| [55] | Zhang X, Cui YN, Yu M, Su BD, Gong W, Balu?ka F, Ko-mis G, ?amaj J, Shan XY, Lin JX (2019). Phosphoryla-tion-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiol 181, 480-498. |