Genome-wide Identification and Comparative Evolutionary Analysis of the R2R3-MYB Transcription Factor Gene Family in Pepper
- 1Plant Genomics Laboratory, College of Life Sciences, Shihezi University, Shihezi 832003, China
2College of Agriculture, Anhui Science and Technology University, Fengyang 233100, China
Received date: 2020-08-10
Accepted date: 2021-02-01
Online published: 2021-02-25
Abstract
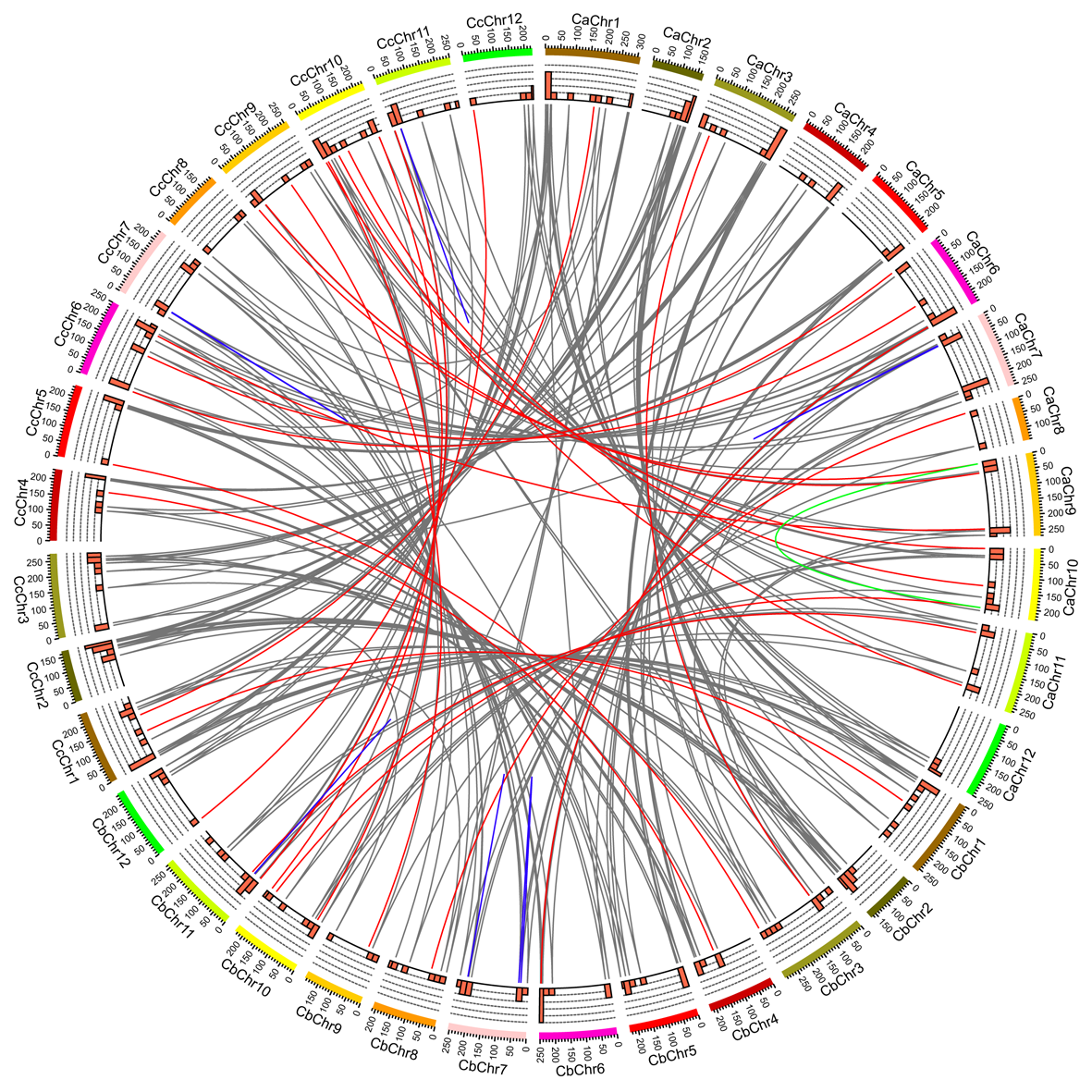
As one of the largest transcription factor (TF) families in plants, MYB TFs are involved in various physiological and biochemical processes, such as plant growth, metabolism, and response to various biotic and abiotic stresses. R2R3-MYB is the main form of MYB TFs in higher plants. Pepper is a vegetable crop with important economic value, but the R2R3-MYB TF family has not been systematically studied in pepper. In this study, 94 CaMYB, 92 CbMYB, and 94 CcMYB TFs genes were identified with comparative genomic analysis in Capsicum annuum, C. baccatum, and C. chinense, respectively. These genes were categorized into 28 subfamilies. Collinearity analysis indicated that there were 73 groups of orthologous R2R3-MYB genes among three pepper species. There were five, four, and two unique R2R3-MYB genes in C. annuum, C. baccatum, and C. chinense, respectively. In addition, we identified 12 pairs of duplicated genes, and eight of which are tandemly repeated genes, which already existed before the divergence of three pepper species. Comparative genomics analysis suggested that the homologous R2R3-MYB TFs underwent functionally divergence during the evolution of pepper. Analysis on the expression profile showed that R2R3-MYB genes were expressed in three major patterns: high expression in roots, leaves, stems, and flowers, such as CaMYB13/CbMYB12/- CcMYB13; high expression in flowers, such as CaMYB93/CbMYB86/CcMYB12; high expression in roots, such as CaMYB48/CbMYB47/CcMYB51. These results lay a foundation for further study on the biological functions of R2R3-MYB TFs in the growth and development of pepper.
Cite this article
Kaicheng Kang , Xiqiang Niu , Xianzhong Huang , Nengbing Hu , Yihu Sui , Kaijing Zhang , Hao Ai . Genome-wide Identification and Comparative Evolutionary Analysis of the R2R3-MYB Transcription Factor Gene Family in Pepper[J]. Chinese Bulletin of Botany, 2021 , 56(3) : 315 -329 . DOI: 10.11983/CBB20143
References
| 1 | 居利香, 雷欣, 赵成志, 舒黄英, 汪志伟, 成善汉 (2020). 辣椒MYB基因家族的鉴定及与辣味关系分析. 园艺学报 47, 875-892. |
| 2 | 李格, 孟小庆, 李宗芸, 朱明库 (2020). 甘薯盐胁迫响应基因IbMYB3的表达特征及生物信息学分析. 植物学报 55, 38-48. |
| 3 | 隋益虎, 陈劲枫 (2009). 辣椒属种间远缘杂交育种研究进展. 热带作物学报 30, 557-563. |
| 4 | 唐嘉瓅, 邱杰, 黄学辉 (2020). 基因组学技术大发展助力园艺植物研究取得新进展. 植物学报 55, 1-4. |
| 5 | 魏家香, 俞佳虹, 程远, 叶青静, 王荣青, 阮美颖, 李志邈, 姚祝平, 周国治, 杨悦俭, 万红建 (2018). 辣椒种间杂交的现状及其研究进展. 分子植物育种 16, 5474-5482. |
| 6 | Blanc G, Wolfe KH (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667-1678. |
| 7 | Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families inArabidopsis thaliana. BMC Plant Biol 4, 10. |
| 8 | Chalhoub B, Denoeud F, Liu SY, Parkin IAP, Tang HB, Wang XY, Chiquet J, Belcram H, Tong CB, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao MX, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier MC, Fan GY, Renault V, Bayer PE, Golicz AA, Manoli S, Lee TH, Thi VHD, Chalabi S, Hu Q, Fan CC, Tollenaere R, Lu YH, Battail C, Shen JX, Sidebottom CHD, Wang XF, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu ZS, Sun FM, Lim YP, Lyons E, Town CD, Bancroft I, Wang XW, Meng JL, Ma JX, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury JM, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou YM, Hua W, Sharpe AG, Paterson AH, Guan CY, Wincker P (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950-953. |
| 9 | Chen SF, Zhou YQ, Chen YR, Gu J (2018). Fastp: an ultra- fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884-i890. |
| 10 | Chen YH, Yang XY, He K, Liu MH, Li JG, Gao ZF, Lin ZQ, Zhang YF, Wang XX, Qiu XM, Shen YP, Zhang L, Deng XH, Luo JC, Deng XW, Chen ZL, Gu HY, Qu LJ (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60, 107-124. |
| 11 | Crooks GE, Hon G, Chandonia JM, Brenner SE (2004). WebLogo: a sequence logo generator. Genome Res 14, 1188-1190. |
| 12 | Dias AP, Braun EL, McMullen MD, Grotewold E (2003). Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol 131, 610-620. |
| 13 | Dossa K, Mmadi MA, Zhou R, Liu AL, Yang YX, Diouf D, You J, Zhang XR (2020). Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis. AoB Plants 12, plz081. |
| 14 | Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX (2012). Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol 12, 106. |
| 15 | Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci 15, 573-581. |
| 16 | Geng P, Zhang S, Liu JY, Zhao CH, Wu J, Cao YP, Fu CX, Han X, He H, Zhao Q (2020). MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol 182, 1272-1283. |
| 17 | Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Muller I, Vo? U, Jürgens G, Ito M (2007). R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134, 1101-1110. |
| 18 | Hajiebrahimi A, Owji H, Hemmati S (2017). Genome-wide identification, functional prediction, and evolutionary analysis of the R2R3-MYB superfamily in Brassica napus. Genome 60, 797-814. |
| 19 | Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R (2015). InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16, 169. |
| 20 | Hu B, Jin JP, Guo AY, Zhang H, Luo JC, Gao G (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296-1297. |
| 21 | Ibiza VP, Blanca J, Ca?izares J, Nuez F (2012). Taxonomy and genetic diversity of domesticated Capsicum species in the Andean region. Genet Resour Crop Evol 59, 1077-1088. |
| 22 | Jia L, Clegg MT, Jiang T (2004). Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol 134, 575-585. |
| 23 | Jiang CZ, Gu JY, Chopra S, Gu X, Peterson T (2004). Ordered origin of the typical two- and three-repeat MYB genes. Gene 326, 13-22. |
| 24 | Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17, 287-291. |
| 25 | Kim S, Park J, Yeom SI, Kim YM, Seo E, Kim KT, Kim MS, Lee JM, Cheong K, Shin HS, Kim SB, Han K, Lee J, Park M, Lee HA, Lee HY, Lee Y, Oh S, Lee JH, Choi E, Choi E, Lee SE, Jeon J, Kim H, Choi G, Song H, Lee J, Lee SC, Kwon JK, Lee HY, Koo N, Hong YJ, Kim RW, Kang WH, Huh JH, Kang BC, Yang TJ, Lee YH, Bennetzen JL, Choi D (2017). New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol 18, 210. |
| 26 | Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009). Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639-1645. |
| 27 | Kumar S, Stecher G, Tamura K (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33, 1870-1874. |
| 28 | Langmead B, Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357-359. |
| 29 | Li B, Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. |
| 30 | Li XX, Guo C, Ahmad S, Wang Q, Yu J, Liu C, Guo YF (2019). Systematic analysis of MYB family genes in potato and their multiple roles in development and stress responses. Biomolecules 9, 317. |
| 31 | Liu YH, Kui LW, Espley RV, Wang L, Li YM, Liu Z, Zhou P, Zeng LH, Zhang XJ, Zhang JL, Allan AC (2019). StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. J Exp Bot 70, 3809-3824. |
| 32 | Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 ?ΔΔCT method . Methods 25, 402-408. |
| 33 | Lotkowska ME, Tohge T, Fernie AR, Xue GP, Balazadeh S, Mueller-Roeber B (2015). The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol 169, 1862-1880. |
| 34 | Matus JT, Aquea F, Arce-Johnson P (2008). Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8, 83. |
| 35 | Ogata K, Kanei-Ishii C, Sasaki M, Hatanaka H, Nagadoi A, Enari M, Nakamura H, Nishimura Y, Ishii S, Sarai A (1996). The cavity in the hydrophobic core of Myb DNA- binding domain is reserved for DNA recognition and trans- activation. Nat Struct Biol 3, 178-187. |
| 36 | Pabo CO, Sauer RT (1992). Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem 61, 1053-1095. |
| 37 | Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J 6, 3553-3558. |
| 38 | Qi XW, Fang HL, Chen ZQ, Liu ZQ, Yu X, Liang CY (2019). Ectopic expression of a R2R3-MYB transcription factor gene LjaMYB12 from Lonicera japonica increases flavonoid accumulation in Arabidopsis thaliana. Int J Mol Sci 20, 4494. |
| 39 | Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu GL (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105-2110. |
| 40 | Rosinski JA, Atchley WR (1998). Molecular evolution of the MYB family of transcription factors: evidence for polyphyletic origin. J Mol Evol 46, 74-83. |
| 41 | Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu SQ, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du JC, Tian ZX, Zhu LC, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoema- ker RC, Jackson SA (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178-183. |
| 42 | Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50, 660-677. |
| 43 | Stracke R, Werber M, Weisshaar B (2001). The R2R3- MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4, 447-456. |
| 44 | Sun WJ, Ma ZT, Chen H, Liu MY (2019). MYB gene family in potato ( Solanum tuberosum L.): genome-wide identification of hormone-responsive reveals their potential functions in growth and development. Int J Mol Sci 20, 4847. |
| 45 | The Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189-195. |
| 46 | The Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635-641. |
| 47 | Ullah A, Ul Qamar MT, Nisar M, Hazrat A, Rahim G, Khan AH, Hayat K, Ahmed S, Ali W, Khan A, Yang XY (2020). Characterization of a novel cotton MYB gene, GhMYB108- like responsive to abiotic stresses. Mol Biol Rep 47, 1573-1581. |
| 48 | Valliyodan B, Nguyen HT (2006). Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol 9, 189-195. |
| 49 | Wang DP, Wan HL, Zhang S, Yu J (2009). γ-MYN: a new algorithm for estimating Ka and Ks with consideration of variable substitution rates. Biol Direct 4, 20. |
| 50 | Wang DP, Zhang YB, Zhang Z, Zhu J, Yu J (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom Proteom Bioinf 8, 77-80. |
| 51 | Wang YC, Liu WJ, Jiang HY, Mao ZL, Wang N, Jiang SH, Xu HF, Yang GX, Zhang ZY, Chen XS (2019). The R2R3-MYB transcription factor MdMYB24-like is involved in methyl jasmonate-induced anthocyanin biosynthesis in apple. Plant Physiol Biochem 139, 273-282. |
| 52 | Wang YP, Tang HB, Debarry JD, Tan X, Li JP, Wang XY, Lee TH, Jin HZ, Marler B, Guo H, Kissinger JC, Paterson AH (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40, e49. |
| 53 | Yadav CB, Bonthala VS, Muthamilarasan M, Pandey G, Khan Y, Prasad M (2015). Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Res 22, 79-90. |
| 54 | Zapata L, Ding J, Willing EM, Hartwig B, Bezdan D, Jiao WB, Patel V, Velikkakam James G, Koornneef M, Ossowski S, Schneeberger K (2016). Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc Natl Acad Sci USA 113, E4052-E4060. |
| 55 | Zhang YL, Zhang CL, Wang GL, Wang YX, Qi CH, Zhao Q, You CX, Li YY, Hao YJ (2019). The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol 19, 362. |
| 56 | Zhang Z, Xiao JF, Wu JY, Zhang HY, Liu GM, Wang XM, Dai L (2012). ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun 419, 779-781. |
| 57 | Zhao PP, Li Q, Li J, Wang LN, Ren ZH (2014). Genome- wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol Genet Genomics 289, 1183-1207. |